| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,400,101 – 10,400,240 |
| Length | 139 |
| Max. P | 0.845536 |

| Location | 10,400,101 – 10,400,207 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.12 |
| Shannon entropy | 0.35851 |
| G+C content | 0.28883 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -11.87 |
| Energy contribution | -12.34 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506743 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
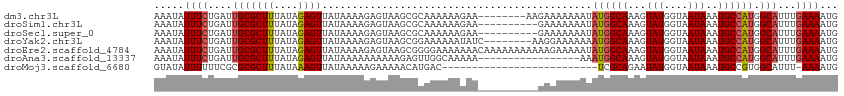
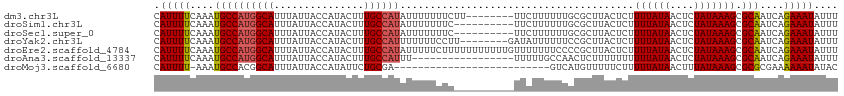
>dm3.chr3L 10400101 106 + 24543557 AAAUAUUUCUGAUUGCGCUUUAUAGAGUUAUAAAAGAGUAAGCGCAAAAAAGAA--------AAGAAAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUG .....(((((..((((((((...................))))))))...))))--------).........(((((((...(((....)))..)))))))............. ( -22.31, z-score = -2.54, R) >droSim1.chr3L 9793469 104 + 22553184 AAAUAUUUCUGAUUGCGCUUUAUAGAGUUAUAAAAGAGUAAGCGCAAAAAAGAA----------GAAAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUG .....(((((..((((((((...................))))))))...))))----------).......(((((((...(((....)))..)))))))............. ( -22.01, z-score = -2.37, R) >droSec1.super_0 2612728 104 + 21120651 AAAUAUUUCUGAUUGCGCUUUAUAGAGUUAUAAAAGAGUAAGCGCAAAAAAGAA----------GAAAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUG .....(((((..((((((((...................))))))))...))))----------).......(((((((...(((....)))..)))))))............. ( -22.01, z-score = -2.37, R) >droYak2.chr3L 10392168 106 + 24197627 AAAUAUUUCUGAUUGCGCUUUAUAGAGUUAUAAAAGAGUAAGCGGAAAAAAUAUC--------AAGGAAAAAAAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUG .....((((((.((((.(((((((....))).)))).)))).)))))).....((--------(((.......((((((...(((....)))..))))))....)))))..... ( -20.60, z-score = -2.00, R) >droEre2.scaffold_4784 10396870 114 + 25762168 AAAUAUUUCUGAUUGCGCUUUAUAGAGUUAUAAAAGAGUAAGCGGGGAAAAAAACAAAAAAAAAAAGAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUG .....(..(((.((((.(((((((....))).)))).)))).)))..)........................(((((((...(((....)))..)))))))............. ( -18.20, z-score = -1.45, R) >droAna3.scaffold_13337 15314077 97 + 23293914 AAAUAUUUCUGAUUGCGCUUUAUAGAGUUAUAAAAAAAAAAGAGUUGGCAAAAA-----------------AAAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUG ...(((((..((.(((((((....))))..........................-----------------..((((((...(((....)))..)))))).))).))..))))) ( -14.50, z-score = 0.01, R) >droMoj3.scaffold_6680 4193551 87 - 24764193 GUAUAUUUUUUCGCGCGCUUUAUAAAGUUAUAAAAAGAAAAACAUGAC--------------------------UCGCAGAAUAUGGUAAUAAAUGCCGUGGCAUUU-AAAAUG ...((((..(((..(((........((((((............)))))--------------------------)))).)))...)))).(((((((....))))))-)..... ( -13.70, z-score = 0.06, R) >consensus AAAUAUUUCUGAUUGCGCUUUAUAGAGUUAUAAAAGAGUAAGCGCAAAAAAGAA__________GAAAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUG ...(((((..((.(((((((....)))).............................................((((((...(((....)))..)))))).))).))..))))) (-11.87 = -12.34 + 0.47)
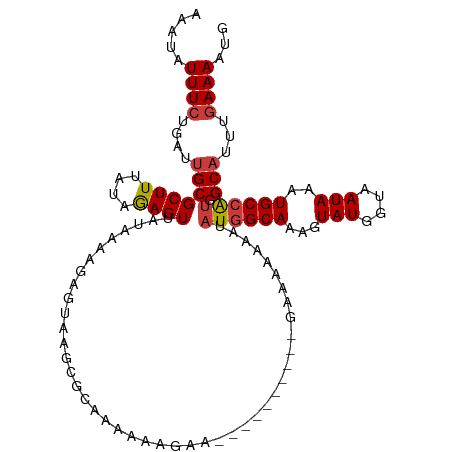
| Location | 10,400,101 – 10,400,207 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.12 |
| Shannon entropy | 0.35851 |
| G+C content | 0.28883 |
| Mean single sequence MFE | -14.57 |
| Consensus MFE | -9.23 |
| Energy contribution | -9.42 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845536 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 10400101 106 - 24543557 CAUUUUCAAAUGCCAUGGCAUUUAUUACCAUACUUUGCCAUAUUUUUUUCUU--------UUCUUUUUUGCGCUUACUCUUUUAUAACUCUAUAAAGCGCAAUCAGAAAUAUUU ..............((((((...............))))))..........(--------((((...((((((((...................))))))))..)))))..... ( -18.77, z-score = -3.44, R) >droSim1.chr3L 9793469 104 - 22553184 CAUUUUCAAAUGCCAUGGCAUUUAUUACCAUACUUUGCCAUAUUUUUUUC----------UUCUUUUUUGCGCUUACUCUUUUAUAACUCUAUAAAGCGCAAUCAGAAAUAUUU ..............((((((...............)))))).........----------((((...((((((((...................))))))))..))))...... ( -17.97, z-score = -3.09, R) >droSec1.super_0 2612728 104 - 21120651 CAUUUUCAAAUGCCAUGGCAUUUAUUACCAUACUUUGCCAUAUUUUUUUC----------UUCUUUUUUGCGCUUACUCUUUUAUAACUCUAUAAAGCGCAAUCAGAAAUAUUU ..............((((((...............)))))).........----------((((...((((((((...................))))))))..))))...... ( -17.97, z-score = -3.09, R) >droYak2.chr3L 10392168 106 - 24197627 CAUUUUCAAAUGCCAUGGCAUUUAUUACCAUACUUUGCCAUUUUUUUCCUU--------GAUAUUUUUUCCGCUUACUCUUUUAUAACUCUAUAAAGCGCAAUCAGAAAUAUUU .......((((((....))))))............................--------(((((((((..(((((...................))))).....))))))))). ( -12.21, z-score = -1.18, R) >droEre2.scaffold_4784 10396870 114 - 25762168 CAUUUUCAAAUGCCAUGGCAUUUAUUACCAUACUUUGCCAUAUUUUUCUUUUUUUUUUUGUUUUUUUCCCCGCUUACUCUUUUAUAACUCUAUAAAGCGCAAUCAGAAAUAUUU ..............((((((...............)))))).................(((((((.....(((((...................))))).....)))))))... ( -11.97, z-score = -1.38, R) >droAna3.scaffold_13337 15314077 97 - 23293914 CAUUUUCAAAUGCCAUGGCAUUUAUUACCAUACUUUGCCAUUU-----------------UUUUUGCCAACUCUUUUUUUUUUAUAACUCUAUAAAGCGCAAUCAGAAAUAUUU ...((((((((((....))))))...........((((((...-----------------....)).............(((((((....))))))).))))...))))..... ( -10.40, z-score = -0.62, R) >droMoj3.scaffold_6680 4193551 87 + 24764193 CAUUUU-AAAUGCCACGGCAUUUAUUACCAUAUUCUGCGA--------------------------GUCAUGUUUUUCUUUUUAUAACUUUAUAAAGCGCGCGAAAAAAUAUAC .....(-((((((....)))))))........((((((((--------------------------(........))).(((((((....))))))).))).)))......... ( -12.70, z-score = -0.61, R) >consensus CAUUUUCAAAUGCCAUGGCAUUUAUUACCAUACUUUGCCAUAUUUUUUUC__________UUCUUUUUUGCGCUUACUCUUUUAUAACUCUAUAAAGCGCAAUCAGAAAUAUUU .(((((....((((((((((...............)))))).......................................((((((....))))))).)))....))))).... ( -9.23 = -9.42 + 0.19)
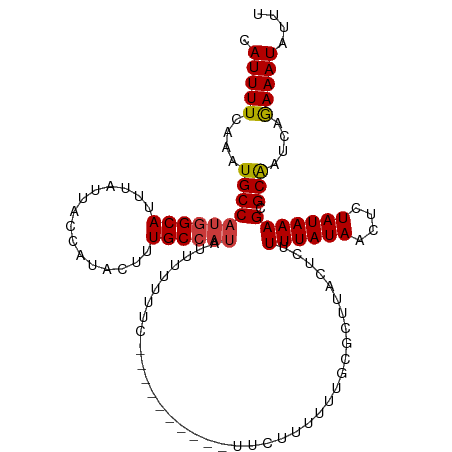
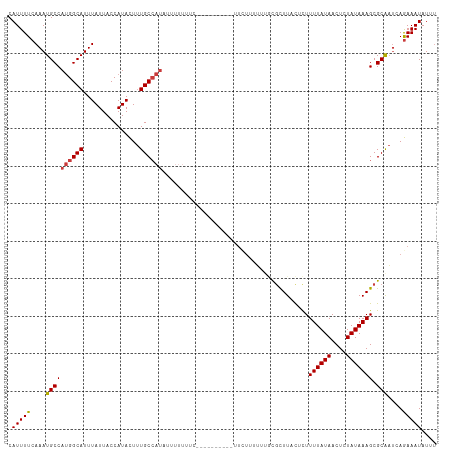
| Location | 10,400,140 – 10,400,240 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Shannon entropy | 0.29352 |
| G+C content | 0.27588 |
| Mean single sequence MFE | -15.70 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651175 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
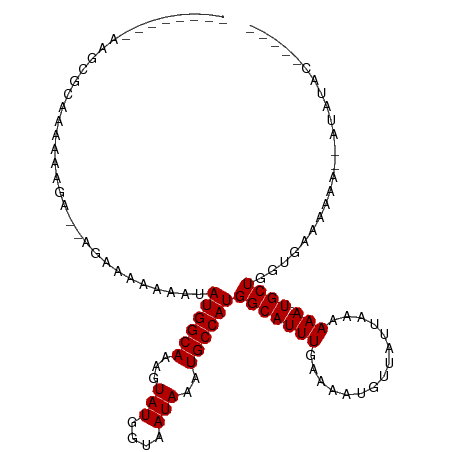
>dm3.chr3L 10400140 100 + 24543557 --------AAGCGCAAAAAAGAAAAGAAAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUGUUAUUAAAAAA-UGCUGGUGAAAAAAAAAUAUAC----- --------...(((....................((((((...(((....)))..))))))(((((((...............)))-)))).)))..............----- ( -15.66, z-score = -1.83, R) >droSim1.chr3L 9793508 96 + 22553184 --------AAGCGCAAAAAAGA--AGAAAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUGUUAUUAAAAAA-UGCUGGUGAAAAAA--AUAUAC----- --------...(((........--..........((((((...(((....)))..))))))(((((((...............)))-)))).)))......--......----- ( -15.66, z-score = -1.64, R) >droSec1.super_0 2612767 96 + 21120651 --------AAGCGCAAAAAAGA--AGAAAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUGUUAUUAAAAAA-UGCUGGUGAAAAAA--AUAUAC----- --------...(((........--..........((((((...(((....)))..))))))(((((((...............)))-)))).)))......--......----- ( -15.66, z-score = -1.64, R) >droYak2.chr3L 10392207 100 + 24197627 --------AAGCGGAAAAAAUAUCAAGGAAAAAAAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUGUUAUUAAAAAAAUGCUGGUGAAAAAAU-AUAUAC----- --------............((((..........((((((...(((....)))..))))))(((((((................))))))))))).......-......----- ( -14.99, z-score = -1.45, R) >droEre2.scaffold_4784 10396909 106 + 25762168 AAGCGGGGAAAAAAACAAAAAAAAAAAGAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUGUUAUUAAAAAA-UGCUGGUGAAAAAA--AUGUAC----- ..(((.........((..................((((((...(((....)))..))))))(((((((...............)))-)))).)).......--.)))..----- ( -14.29, z-score = -1.35, R) >droAna3.scaffold_13337 15314112 100 + 23293914 -------------AAAAAAGAGUUGGCAAAAAAAAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUGUUAUGAAAAAA-UGCUGCUGAAGAAAAAAGAAACAAGCC -------------.........(..((.......((((((...(((....)))..))))))(((((((...............)))-))))))..).................. ( -17.96, z-score = -1.80, R) >consensus ________AAGCGCAAAAAAGA__AGAAAAAAAUAUGGCAAAGUAUGGUAAUAAAUGCCAUGGCAUUUGAAAAUGUUAUUAAAAAA_UGCUGGUGAAAAAA__AUAUAC_____ ....................................((((...(((((((.....)))))))(((((....)))))...........))))....................... (-11.25 = -11.25 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:10 2011