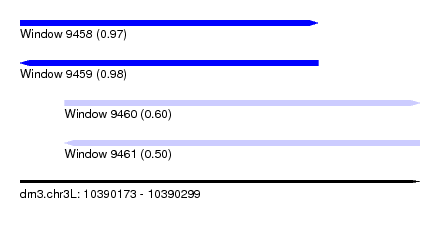
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,390,173 – 10,390,299 |
| Length | 126 |
| Max. P | 0.979205 |

| Location | 10,390,173 – 10,390,267 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.15 |
| Shannon entropy | 0.38375 |
| G+C content | 0.45748 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -19.87 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10390173 94 + 24543557 ------------GCCACAACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUACUGGCCAUCC--AGCGUCCCGAGGCAUUGAUUAAAAUG ------------(((.........(((((((((((((....)).)))))))).)))..............((((....))--))........)))............. ( -26.30, z-score = -1.42, R) >droSim1.chr3L 9783135 94 + 22553184 ------------GCCACAACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCC--AGCGUCCCGAGGCAUUGAUUAAAAUG ------------.........((((((((((((((((....)).)))))))).)))..((((((...(((((((....))--))))).....)))))))))....... ( -32.00, z-score = -2.75, R) >droSec1.super_0 2602805 94 + 21120651 ------------GCCACAACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCC--AGCGUCCCGAGGCAUUGAUUAAAAUG ------------.........((((((((((((((((....)).)))))))).)))..((((((...(((((((....))--))))).....)))))))))....... ( -32.00, z-score = -2.75, R) >droYak2.chr3L 10382085 94 + 24197627 ------------GCCACAACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCC--AGCGUCCCGAGGCAUUGAUUAAAAUG ------------.........((((((((((((((((....)).)))))))).)))..((((((...(((((((....))--))))).....)))))))))....... ( -32.00, z-score = -2.75, R) >droEre2.scaffold_4784 10386531 96 + 25762168 ------------GCCACAACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCUCGAGCGUCCCGAGGCAUUGAUUAAAAUG ------------(((......((((((((((((((((....)).)))))))).((......))......))))))....(((.......))))))............. ( -29.90, z-score = -1.96, R) >droAna3.scaffold_13337 15304421 91 + 23293914 ------------GCUGAAACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCCGGCCAUCC-----ACCAAGAGGCAUUGAUUAAAAUU ------------(((((.....)))))((((((((((....)).)))))))).((......))...((((((........-----.......)))))).......... ( -26.86, z-score = -1.96, R) >droWil1.scaffold_181024 1582606 84 - 1751709 ---------UUUGCUUUAGCUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGUUAAA--------CAUCU-------UGCAAGGCAUUGAUUAAAAUU ---------..((((((.((....(((((((((((((....)).)))))))).)))...(((.....--------)))..-------.))))))))............ ( -24.40, z-score = -1.90, R) >droMoj3.scaffold_6680 4181444 93 - 24764193 GCCACUAGCUGUUGCCUGUCUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUUAAAUGUAAAA--------CAUUC-------AGCGAAGCAUUGAUUAAAAUU (((((..((((((..(.((......)).)..)))))).)))))..((((..((((((.((((.....--------)))).-------....))))))))))....... ( -25.40, z-score = -1.10, R) >droVir3.scaffold_13049 16176411 93 + 25233164 ACCUCUAGCUGUUGCCUGUCUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGUAAAA--------CAUUC-------AACGAGGCAUUGAUUAAAAUU .((((((((((((((.((((...(.((((....)))).).)))).).))))).)))).((((.....--------)))).-------...)))).............. ( -25.50, z-score = -1.83, R) >consensus ____________GCCACAACUGUCAGCUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCC__AGCGUCCCGAGGCAUUGAUUAAAAUG .....................((((((((((((((((....)).)))))))).)))..((((((............................)))))))))....... (-19.87 = -19.87 + 0.00)

| Location | 10,390,173 – 10,390,267 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.15 |
| Shannon entropy | 0.38375 |
| G+C content | 0.45748 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -18.93 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10390173 94 - 24543557 CAUUUUAAUCAAUGCCUCGGGACGCU--GGAUGGCCAGUAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUUGUGGC------------ (((.((((..(((((...((((..((--((....))))..)))).))))))))).))).......(((((((((((((.....)))))))))))))------------ ( -33.20, z-score = -2.51, R) >droSim1.chr3L 9783135 94 - 22553184 CAUUUUAAUCAAUGCCUCGGGACGCU--GGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUUGUGGC------------ (((.((((..(((((...((((.(((--((....))))).)))).))))))))).))).......(((((((((((((.....)))))))))))))------------ ( -34.20, z-score = -2.56, R) >droSec1.super_0 2602805 94 - 21120651 CAUUUUAAUCAAUGCCUCGGGACGCU--GGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUUGUGGC------------ (((.((((..(((((...((((.(((--((....))))).)))).))))))))).))).......(((((((((((((.....)))))))))))))------------ ( -34.20, z-score = -2.56, R) >droYak2.chr3L 10382085 94 - 24197627 CAUUUUAAUCAAUGCCUCGGGACGCU--GGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUUGUGGC------------ (((.((((..(((((...((((.(((--((....))))).)))).))))))))).))).......(((((((((((((.....)))))))))))))------------ ( -34.20, z-score = -2.56, R) >droEre2.scaffold_4784 10386531 96 - 25762168 CAUUUUAAUCAAUGCCUCGGGACGCUCGAGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUUGUGGC------------ ...............((((((...))))))(((..((((((.((((....)))).)))))).)))(((((((((((((.....)))))))))))))------------ ( -34.40, z-score = -2.78, R) >droAna3.scaffold_13337 15304421 91 - 23293914 AAUUUUAAUCAAUGCCUCUUGGU-----GGAUGGCCGGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUUUCAGC------------ .............(((....)))-----.((((((((((((.(((((((......))))))).)))))..)))))))....(((((.....)))))------------ ( -30.10, z-score = -2.33, R) >droWil1.scaffold_181024 1582606 84 + 1751709 AAUUUUAAUCAAUGCCUUGCA-------AGAUG--------UUUAACAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGCUAAAGCAAA--------- ............(((.((((.-------.((((--------.....))))(((((.((((((((.((....)))))))))))))))..)).)).)))..--------- ( -24.90, z-score = -2.75, R) >droMoj3.scaffold_6680 4181444 93 + 24764193 AAUUUUAAUCAAUGCUUCGCU-------GAAUG--------UUUUACAUUUAAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGACAGGCAACAGCUAGUGGC ............(((((..((-------(((((--------.....))))..(((.((((((((.((....)))))))))))))..)))..)))))............ ( -26.00, z-score = -1.25, R) >droVir3.scaffold_13049 16176411 93 - 25233164 AAUUUUAAUCAAUGCCUCGUU-------GAAUG--------UUUUACAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGACAGGCAACAGCUAGAGGU .............((((((((-------(..((--------(((......(((((.((((((((.((....))))))))))))))).)))))..)))).....))))) ( -30.90, z-score = -3.41, R) >consensus CAUUUUAAUCAAUGCCUCGGGACGCU__GGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAGCUGACAGUUGUGGC____________ ..................................................(((((.((((((((.((....)))))))))))))))...................... (-18.93 = -19.04 + 0.11)
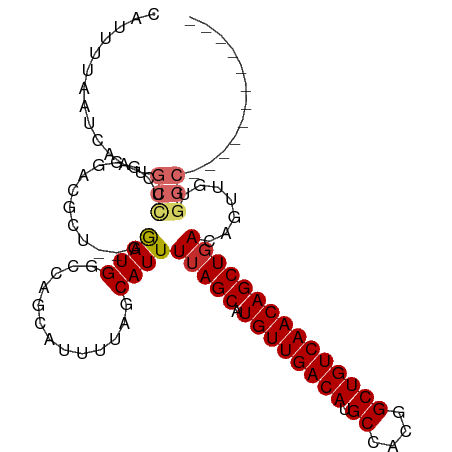
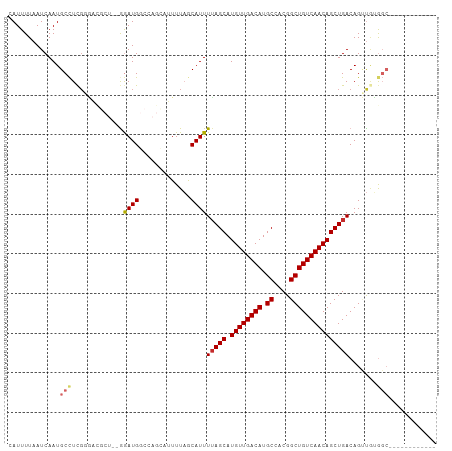
| Location | 10,390,187 – 10,390,299 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.26 |
| Shannon entropy | 0.40298 |
| G+C content | 0.42307 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10390187 112 + 24543557 CUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUACUGGCCAUCC--AGCGUCCCGAGGCAUUGAUUAAAAUGGCUUCUUUUUUUUCCUCAUCUCAUCUGAGCUU------ ..(((((((((....)).)))))))(((((.....((((......)))).....--)))))...((((((((......))).))))).........((((.......))))...------ ( -27.20, z-score = -1.01, R) >droSim1.chr3L 9783149 110 + 22553184 CUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCC--AGCGUCCCGAGGCAUUGAUUAAAAUGGCUUCUUUUUUU--UUCCUCUCAUCUGCGCUU------ .....((.((((((((((((....))))))..((((((...(((((((....))--))))).....)))))).......)))))))).......--..................------ ( -29.70, z-score = -1.36, R) >droSec1.super_0 2602819 110 + 21120651 CUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCC--AGCGUCCCGAGGCAUUGAUUAAAAUGGCUUCUUUUUUU--CUCCUCUCAUCUGCGCUU------ .....((.((((((((((((....))))))..((((((...(((((((....))--))))).....)))))).......)))))))).......--..................------ ( -29.70, z-score = -1.45, R) >droEre2.scaffold_4784 10386545 112 + 25762168 CUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCUCGAGCGUCCCGAGGCAUUGAUUAAAAUGGCUUAUUUUUUU--UCUCUCUCAUCUGCGCUU------ .((((((((((....)).)))))))).((((....((......))))))......((((((...((((....((..((((........))))..--))..))))....))))))------ ( -27.30, z-score = -0.46, R) >droAna3.scaffold_13337 15304435 109 + 23293914 CUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCCGGCCAUCC-----ACCAAGAGGCAUUGAUUAAAAUUUUUCCUUUUUUUGUUUCAAGCCUUGCUAGCCU------ .((((((((((....)).)))))))).((......))......((..((.....-----.....(((((.((((.(((((..........)))))..)))))))))))..))..------ ( -27.50, z-score = -1.21, R) >droVir3.scaffold_13049 16176437 105 + 25233164 CUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGUAAAA--------CAUUC-------AACGAGGCAUUGAUUAAAAUUGUUUUCCUAUACAAAUUAAAUAAAUAAAGUGUUAGCAU .(((((((((((.(((((((....))))))).((((.....--------)))).-------.....)))...........((((........))))...............)))))))). ( -23.70, z-score = -2.22, R) >consensus CUGUUGACAGCCGUGGCAUGUCAACAUGCUAAAAUGCUAAAAUGCUGGCCAUCC__AGCGUCCCGAGGCAUUGAUUAAAAUGGCUUCUUUUUUU__UUCAUCUCAUCUGAGCUU______ .((((((((((....)).)))))))).((......((((......))))...............(((.((((......)))).)))........................))........ (-17.90 = -18.57 + 0.67)

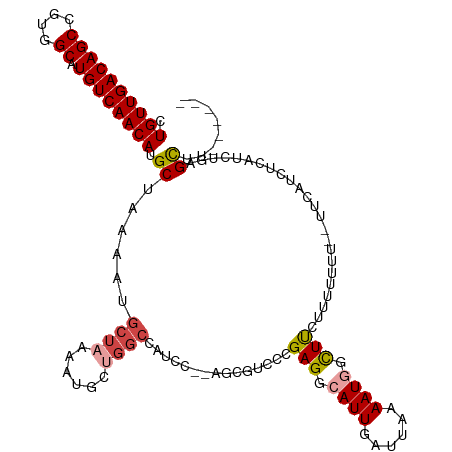
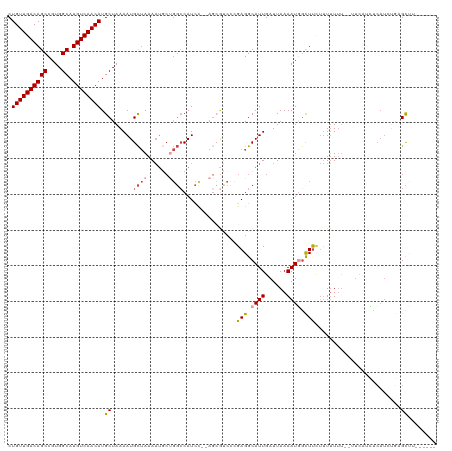
| Location | 10,390,187 – 10,390,299 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.26 |
| Shannon entropy | 0.40298 |
| G+C content | 0.42307 |
| Mean single sequence MFE | -30.21 |
| Consensus MFE | -15.78 |
| Energy contribution | -17.06 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 10390187 112 - 24543557 ------AAGCUCAGAUGAGAUGAGGAAAAAAAAGAAGCCAUUUUAAUCAAUGCCUCGGGACGCU--GGAUGGCCAGUAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAG ------..(((.(((((....((((.......((((....))))........))))((((..((--((....))))..))))..))))).))).((((((((.((....)))))))))). ( -33.16, z-score = -2.06, R) >droSim1.chr3L 9783149 110 - 22553184 ------AAGCGCAGAUGAGAGGAA--AAAAAAAGAAGCCAUUUUAAUCAAUGCCUCGGGACGCU--GGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAG ------..((..(((((.((((..--......((((....))))........)))).....(((--((....))))).......)))))..)).((((((((.((....)))))))))). ( -32.69, z-score = -1.81, R) >droSec1.super_0 2602819 110 - 21120651 ------AAGCGCAGAUGAGAGGAG--AAAAAAAGAAGCCAUUUUAAUCAAUGCCUCGGGACGCU--GGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAG ------..((..(((((.((((..--......((((....))))........)))).....(((--((....))))).......)))))..)).((((((((.((....)))))))))). ( -32.69, z-score = -1.75, R) >droEre2.scaffold_4784 10386545 112 - 25762168 ------AAGCGCAGAUGAGAGAGA--AAAAAAAUAAGCCAUUUUAAUCAAUGCCUCGGGACGCUCGAGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAG ------....((............--..........(((((............((((((...)))))))))))..((......))......)).((((((((.((....)))))))))). ( -27.80, z-score = -0.64, R) >droAna3.scaffold_13337 15304435 109 - 23293914 ------AGGCUAGCAAGGCUUGAAACAAAAAAAGGAAAAAUUUUAAUCAAUGCCUCUUGGU-----GGAUGGCCGGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAG ------..(((((..((((((((.......................)))).)))).)))))-----.((((((((((((.(((((((......))))))).)))))..)))))))..... ( -31.20, z-score = -1.34, R) >droVir3.scaffold_13049 16176437 105 - 25233164 AUGCUAACACUUUAUUUAUUUAAUUUGUAUAGGAAAACAAUUUUAAUCAAUGCCUCGUU-------GAAUG--------UUUUACAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAG .((((((((..(((......)))..)))...(.((((((.......((((((...))))-------)).))--------)))).)....)))))((((((((.((....)))))))))). ( -23.70, z-score = -2.49, R) >consensus ______AAGCGCAGAUGAGAGGAA__AAAAAAAGAAGCCAUUUUAAUCAAUGCCUCGGGACGCU__GGAUGGCCAGCAUUUUAGCAUUUUAGCAUGUUGACAUGCCACGGCUGUCAACAG ........((..........................(((((((.........(.....).......)))))))..((......))......)).((((((((.((....)))))))))). (-15.78 = -17.06 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:08 2011