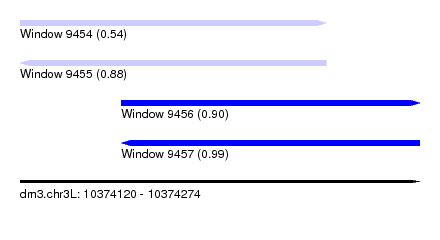
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,374,120 – 10,374,274 |
| Length | 154 |
| Max. P | 0.991846 |

| Location | 10,374,120 – 10,374,238 |
|---|---|
| Length | 118 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.72 |
| Shannon entropy | 0.17706 |
| G+C content | 0.44443 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -28.33 |
| Energy contribution | -28.37 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
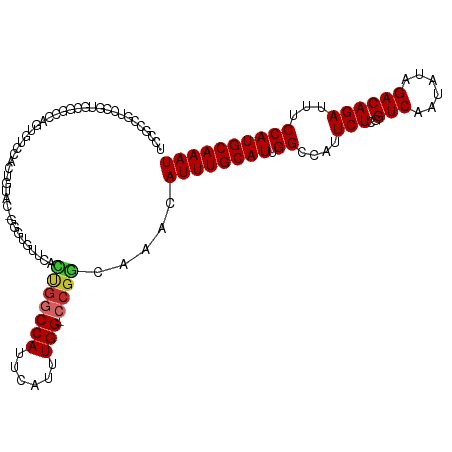
>dm3.chr3L 10374120 118 + 24543557 AAUUUGGCUAGGGCUCGGCCC-GAAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACA ...(((((((...(((((..(-((.......(((.........)))(((((......))))).))).)))))..)))))))......((((..(.((-(((......))))).)..)))) ( -38.20, z-score = -2.05, R) >droAna3.scaffold_13337 15287548 118 + 23293914 AAUUUGGCUAGGGCCUGGCCC-GAAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGACAGUCGUCC .((((((((.((((...))))-.(((((((((((((.......))).(((...((.(((......))).))......)))..))))))))))...))-))))))....((((....)))) ( -37.00, z-score = -1.96, R) >droEre2.scaffold_4784 10370090 119 + 25762168 AAUUUGGCUAGGGCUCGGCCCCGAAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACA ...(((((((...(((((.............(((.........)))(((((......))))).....)))))..)))))))......((((..(.((-(((......))))).)..)))) ( -37.50, z-score = -1.64, R) >droYak2.chr3L 10365408 118 + 24197627 AAUUUGGCUAGGGCCCGGCCC-GAAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACA ..........((((...))))-....((((..((((....((((((((((...((.(((......))).))......)).))))))))...))).((-(((......))))).)..)))) ( -37.70, z-score = -1.72, R) >droSec1.super_0 2586885 118 + 21120651 AAUUUGGCUAGGGCUCGGCCC-GAAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACA ...(((((((...(((((..(-((.......(((.........)))(((((......))))).))).)))))..)))))))......((((..(.((-(((......))))).)..)))) ( -38.20, z-score = -2.05, R) >droSim1.chr3L 9766577 118 + 22553184 AAUUUGGCUAGGGCUCGGCCC-GAAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACA ...(((((((...(((((..(-((.......(((.........)))(((((......))))).))).)))))..)))))))......((((..(.((-(((......))))).)..)))) ( -38.20, z-score = -2.05, R) >droWil1.scaffold_181024 1559576 107 - 1751709 AAUUUGGCUAGGGCC-GGCCC--AAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCUGG-CCAAAUGAAUGGG--------- .((((((((((((..-..))(--(((((((((((((.......))).(((...((.(((......))).))......)))..)))))))))))))))-)))))).......--------- ( -34.20, z-score = -1.90, R) >droVir3.scaffold_13049 16160100 116 + 25233164 AAUUUGACUAGGGCC--GCCC-CGAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGUCGGGCCAAAUGAAUGGCCGAGAACG- .......((..((((--((((-((((((((((((((.......))).(((...((.(((......))).))......)))..)))))))))))..))))(....)...)))).))....- ( -33.10, z-score = -1.44, R) >droMoj3.scaffold_6680 4162413 115 - 24764193 AAUUUGACUAGGGCC--GCCC--GAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGGGCCAAAUGAAUGGCCGAGAAUG- .......((..((((--((((--(((((((((((((.......))).(((...((.(((......))).))......)))..)))))))))...)))))(....)...)))).))....- ( -31.90, z-score = -1.21, R) >droGri2.scaffold_15110 12059708 116 - 24565398 AAUUUGGAUAGGGCC--GCCC-CGAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGGGCCAAAUGAAUGGCCGAGAACG- ...........((((--((((-((((((((((((((.......))).(((...((.(((......))).))......)))..)))))))))))..))))(....)...)))).......- ( -32.70, z-score = -0.81, R) >consensus AAUUUGGCUAGGGCCCGGCCC_GAAAUGUUUGCGUAUUAAAUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG_CCAAAUGAAUGGCCAGUGAACA .....(((..((((...))))..(((((((((((((.......))).(((...((.(((......))).))......)))..)))))))))))))((.(((......)))))........ (-28.33 = -28.37 + 0.04)

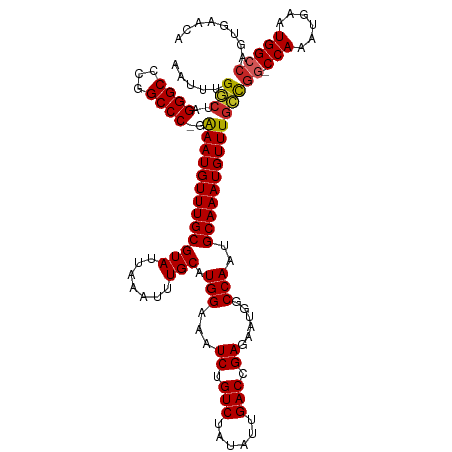
| Location | 10,374,120 – 10,374,238 |
|---|---|
| Length | 118 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.72 |
| Shannon entropy | 0.17706 |
| G+C content | 0.44443 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -26.84 |
| Energy contribution | -27.09 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10374120 118 - 24543557 UGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUUC-GGGCCGAGCCCUAGCCAAAUU ........(((((......)))-))(((.....((((((((((((......)))))))......((((((......)))))).....)))))......-((((...))))..)))..... ( -34.30, z-score = -2.51, R) >droAna3.scaffold_13337 15287548 118 - 23293914 GGACGACUGUCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUUC-GGGCCAGGCCCUAGCCAAAUU ((((....)))).......(((-((.(.(((..((((((((((((......)))))))......((((((......)))))).....)))))..))))-.)))))(((....)))..... ( -33.90, z-score = -2.45, R) >droEre2.scaffold_4784 10370090 119 - 25762168 UGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUUCGGGGCCGAGCCCUAGCCAAAUU ........(((((......)))-))(((.....((((((((((((......)))))))......((((((......)))))).....)))))......(((((...))))).)))..... ( -35.40, z-score = -2.38, R) >droYak2.chr3L 10365408 118 - 24197627 UGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUUC-GGGCCGGGCCCUAGCCAAAUU ........(((((......)))-))(((.....((((((((((((......)))))))......((((((......)))))).....)))))......-((((...))))..)))..... ( -34.30, z-score = -1.94, R) >droSec1.super_0 2586885 118 - 21120651 UGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUUC-GGGCCGAGCCCUAGCCAAAUU ........(((((......)))-))(((.....((((((((((((......)))))))......((((((......)))))).....)))))......-((((...))))..)))..... ( -34.30, z-score = -2.51, R) >droSim1.chr3L 9766577 118 - 22553184 UGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUUC-GGGCCGAGCCCUAGCCAAAUU ........(((((......)))-))(((.....((((((((((((......)))))))......((((((......)))))).....)))))......-((((...))))..)))..... ( -34.30, z-score = -2.51, R) >droWil1.scaffold_181024 1559576 107 + 1751709 ---------CCCAUUCAUUUGG-CCAGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUU--GGGCC-GGCCCUAGCCAAAUU ---------...........((-((..((((..((((((((((((......)))))))......((((((......)))))).....)))))..)))--)))))-(((....)))..... ( -27.30, z-score = -1.65, R) >droVir3.scaffold_13049 16160100 116 - 25233164 -CGUUCUCGGCCAUUCAUUUGGCCCGACAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUCG-GGGC--GGCCCUAGUCAAAUU -.......(((((......))))).(((.....((((((((((((......)))))))......((((((......)))))).....))))).....(-((..--..)))..)))..... ( -29.70, z-score = -1.54, R) >droMoj3.scaffold_6680 4162413 115 + 24764193 -CAUUCUCGGCCAUUCAUUUGGCCCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUC--GGGC--GGCCCUAGUCAAAUU -.......(((((......))))).(((....((((((((.((....(((..(((......))))))...))))))))))......(((...(....--).))--))))........... ( -29.10, z-score = -1.60, R) >droGri2.scaffold_15110 12059708 116 + 24565398 -CGUUCUCGGCCAUUCAUUUGGCCCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUCG-GGGC--GGCCCUAUCCAAAUU -.......(((((......))))).(((....((((((((.((....(((..(((......))))))...))))))))))......(((...(....)-..))--))))........... ( -29.50, z-score = -1.04, R) >consensus UGUUCACUGGCCAUUCAUUUGG_CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAUUUAAUACGCAAACAUUUC_GGGCCGGGCCCUAGCCAAAUU ........(((((......))).))(((.....((((((((((((......)))))))......((((((......)))))).....))))).......((((...))))..)))..... (-26.84 = -27.09 + 0.25)

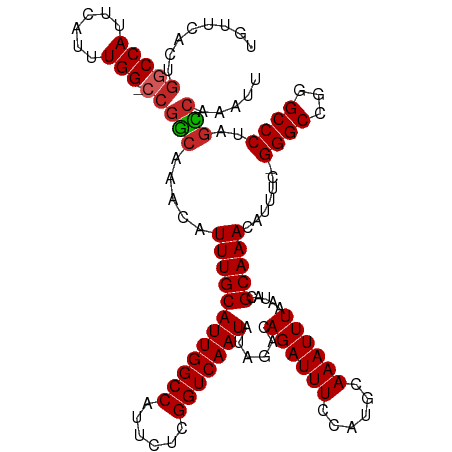
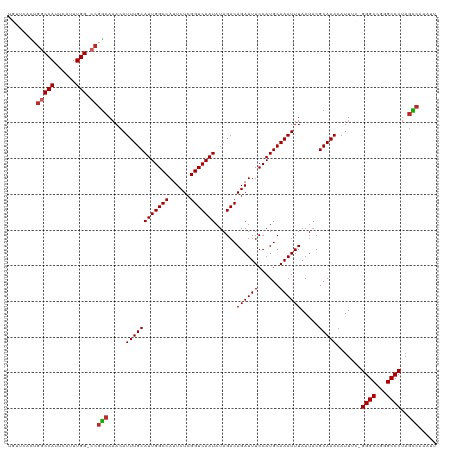
| Location | 10,374,159 – 10,374,274 |
|---|---|
| Length | 115 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Shannon entropy | 0.40726 |
| G+C content | 0.49108 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -18.21 |
| Energy contribution | -18.39 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10374159 115 + 24543557 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACACCCCGUACAGUGGACACUGGCCGCACGGACAGCGGA .((((((((((......)))))..((.(((....(((((...((((....)))).))-))).......((((((((..(((........)))..))))))))...))).))))))) ( -41.70, z-score = -2.60, R) >droAna3.scaffold_13337 15287587 114 + 23293914 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGACAGUCGUCCACC-GUACAGUGGACAUUGGUGCAAUGGCCAAUGGC ..((((((.((...((((((((.((....))...(((((...((((....)))).))-))).....))))))))..((((((.-.....)))))).)).))))))..(((...))) ( -36.90, z-score = -1.36, R) >droEre2.scaffold_4784 10370130 114 + 25762168 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACACCC-GUACAGUGGACACUGGCCGCACGGACGGCGGA ((((((((((...((.(((......))).))......)).)))))))).(((((((.-((...((...((((((((..(((..-.....)))..)))))))).)).)).))))))) ( -44.60, z-score = -3.26, R) >droYak2.chr3L 10365447 114 + 24197627 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACACCC-GUACAGUGGACACUGGCCACACGGACGGCGGA ((((((((((...((.(((......))).))......)).)))))))).(((((((.-((...((..(((((((((..(((..-.....)))..))))))))))).)).))))))) ( -45.00, z-score = -3.67, R) >droSec1.super_0 2586924 114 + 21120651 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACACCC-GUACAGUGGACACUGGCCGCACGGACGGCGGA ((((((((((...((.(((......))).))......)).)))))))).(((((((.-((...((...((((((((..(((..-.....)))..)))))))).)).)).))))))) ( -44.60, z-score = -3.26, R) >droSim1.chr3L 9766616 114 + 22553184 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG-CCAAAUGAAUGGCCAGUGAACACCC-GUACAGUGGACACUGGCCGCACGGACGGCGGA ((((((((((...((.(((......))).))......)).)))))))).(((((((.-((...((...((((((((..(((..-.....)))..)))))))).)).)).))))))) ( -44.60, z-score = -3.26, R) >droWil1.scaffold_181024 1559613 107 - 1751709 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCUGG-CCAAAUGAAUGGGUC-CAAGCGGCA--GUCAACGGCCAGCAACAGAAAGGGAA----- ((((((((((...((.(((......))).))......)).))))))))..(((((((-((...(((....(((-.....))).--.)))..)))))))))...........----- ( -34.30, z-score = -1.62, R) >droVir3.scaffold_13049 16160137 94 + 25233164 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGUCGGGCCAAAUGAAUGGCCGAGAACGAAGA--GACAGCGGCAA-------------------- ((((((((((...((.(((......))).))......)).))))))))..(((((((((((......))))).....(.....--)....))))))-------------------- ( -27.10, z-score = -1.63, R) >droMoj3.scaffold_6680 4162449 94 - 24764193 AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGGGCCAAAUGAAUGGCCGAGAAUGAAGA--GACAGCAGCAA-------------------- .(((((((((...((.(((......))).))......)).)))))))((((.((..(((((......))))).....((....--..)))))))).-------------------- ( -24.30, z-score = -0.89, R) >consensus AUUUGCAUGGAAAUCUGUCUAUAUUGACCGAGAAUGGCCAAUGCAAAUGUUUGCCGG_CCAAAUGAAUGGCCAGUGAACACCC_GUACAGUGGACACUGGCCGCACGGACGGCGGA ((((((((((...((.(((......))).))......)).))))))))..((((.((.(((......))))).))))....................................... (-18.21 = -18.39 + 0.18)

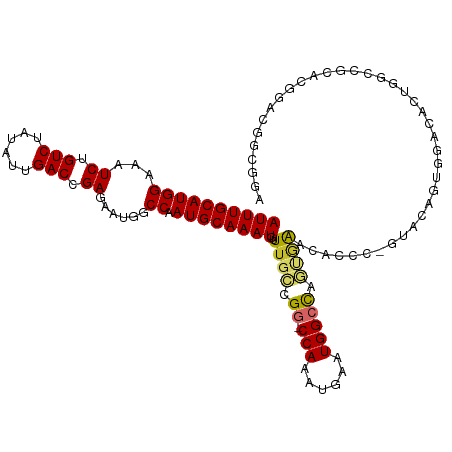
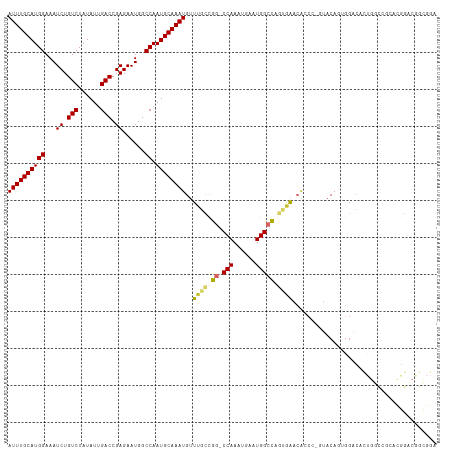
| Location | 10,374,159 – 10,374,274 |
|---|---|
| Length | 115 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Shannon entropy | 0.40726 |
| G+C content | 0.49108 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10374159 115 - 24543557 UCCGCUGUCCGUGCGGCCAGUGUCCACUGUACGGGGUGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU ...((((.(((((.((((((((..((((......))))..))))))))...)))..))-.))))....((((((((.((....(((..(((......))))))...)))))))))) ( -42.00, z-score = -2.51, R) >droAna3.scaffold_13337 15287587 114 - 23293914 GCCAUUGGCCAUUGCACCAAUGUCCACUGUAC-GGUGGACGACUGUCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU ((...((((((.((((.....(((((((....-)))))))(.(((.(((......)))-.)))).......)))).))))))...((.(((......))).)).......)).... ( -38.10, z-score = -2.66, R) >droEre2.scaffold_4784 10370130 114 - 25762168 UCCGCCGUCCGUGCGGCCAGUGUCCACUGUAC-GGGUGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU ...((((.(((((.((((((((..((((....-.))))..))))))))...)))..))-.))))....((((((((.((....(((..(((......))))))...)))))))))) ( -44.00, z-score = -3.48, R) >droYak2.chr3L 10365447 114 - 24197627 UCCGCCGUCCGUGUGGCCAGUGUCCACUGUAC-GGGUGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU ...((((.(((.((((((((((..((((....-.))))..)))))))))).....)))-.))))....((((((((.((....(((..(((......))))))...)))))))))) ( -47.40, z-score = -4.53, R) >droSec1.super_0 2586924 114 - 21120651 UCCGCCGUCCGUGCGGCCAGUGUCCACUGUAC-GGGUGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU ...((((.(((((.((((((((..((((....-.))))..))))))))...)))..))-.))))....((((((((.((....(((..(((......))))))...)))))))))) ( -44.00, z-score = -3.48, R) >droSim1.chr3L 9766616 114 - 22553184 UCCGCCGUCCGUGCGGCCAGUGUCCACUGUAC-GGGUGUUCACUGGCCAUUCAUUUGG-CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU ...((((.(((((.((((((((..((((....-.))))..))))))))...)))..))-.))))....((((((((.((....(((..(((......))))))...)))))))))) ( -44.00, z-score = -3.48, R) >droWil1.scaffold_181024 1559613 107 + 1751709 -----UUCCCUUUCUGUUGCUGGCCGUUGAC--UGCCGCUUG-GACCCAUUCAUUUGG-CCAGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU -----.......((((((((((((((.(((.--((((....)-)...)).)))..)))-)))))..........(((((((......)))))))...))))))............. ( -32.80, z-score = -2.46, R) >droVir3.scaffold_13049 16160137 94 - 25233164 --------------------UUGCCGCUGUC--UCUUCGUUCUCGGCCAUUCAUUUGGCCCGACAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU --------------------((((..(((((--(....(((.(((((((......))).))))..)))......(((((((......)))))))..))))))........)))).. ( -26.60, z-score = -2.83, R) >droMoj3.scaffold_6680 4162449 94 + 24764193 --------------------UUGCUGCUGUC--UCUUCAUUCUCGGCCAUUCAUUUGGCCCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU --------------------(((((((((((--(..........(((((......)))))..((((....))))(((((((......)))))))..)))))).....)).)))).. ( -28.50, z-score = -3.04, R) >consensus UCCGCCGUCCGUGCGGCCAGUGUCCACUGUAC_GGGUGUUCACUGGCCAUUCAUUUGG_CCGGCAAACAUUUGCAUUGGCCAUUCUCGGUCAAUAUAGACAGAUUUCCAUGCAAAU ..........................................(((((((......))).)))).....((((((((.((....(((..(((......))))))...)))))))))) (-18.40 = -18.58 + 0.17)
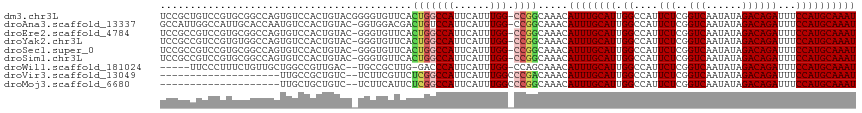
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:16:05 2011