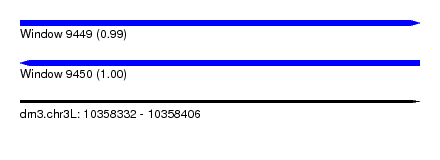
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,358,332 – 10,358,406 |
| Length | 74 |
| Max. P | 0.998488 |

| Location | 10,358,332 – 10,358,406 |
|---|---|
| Length | 74 |
| Sequences | 15 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 85.48 |
| Shannon entropy | 0.31396 |
| G+C content | 0.42503 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -30.19 |
| Energy contribution | -30.41 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -6.44 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10358332 74 + 24543557 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUU--CAUUAUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((..--.........--------..))))))))))..))))).))))..))). ( -32.32, z-score = -6.96, R) >droGri2.scaffold_15110 12043374 76 - 24565398 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUAAAUUUAAAUA--------CUCAUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((.(.............--------..).))))))))..))))).))))..))). ( -26.46, z-score = -5.20, R) >droMoj3.scaffold_6680 4142271 76 - 24764193 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAAUUUAAAUA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((.............--------..))))))))))..))))).))))..))). ( -32.16, z-score = -6.90, R) >droVir3.scaffold_13049 16143990 76 + 25233164 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAAUUUAUAUA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((.............--------..))))))))))..))))).))))..))). ( -32.16, z-score = -6.93, R) >droWil1.scaffold_181024 1528994 76 - 1751709 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAUUUAAUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((.............--------..))))))))))..))))).))))..))). ( -32.16, z-score = -6.90, R) >droPer1.super_27 1191224 76 + 1201981 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAUAUUAUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((.............--------..))))))))))..))))).))))..))). ( -32.16, z-score = -6.85, R) >dp4.chrXR_group6 8756412 76 + 13314419 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAUAUUAUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((.............--------..))))))))))..))))).))))..))). ( -32.16, z-score = -6.85, R) >droAna3.scaffold_13337 15274257 76 + 23293914 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUAACAUUAUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((.............--------..))))))))))..))))).))))..))). ( -32.16, z-score = -6.82, R) >droEre2.scaffold_4784 10353140 74 + 25762168 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUU--CAUUAUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((..--.........--------..))))))))))..))))).))))..))). ( -32.32, z-score = -6.96, R) >droYak2.chr3L 10348890 74 + 24197627 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAU--UAUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((.....--......--------..))))))))))..))))).))))..))). ( -32.32, z-score = -6.96, R) >droSec1.super_0 2571170 74 + 21120651 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAU--UAUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((.....--......--------..))))))))))..))))).))))..))). ( -32.32, z-score = -6.96, R) >droSim1.chr3L 9747999 74 + 22553184 -----UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAUU--AUAAA--------CUCGUAGGAACUUCAUACCGUGCUCUUGGA -----......(((..((((.(((((..((((((((((......--.....--------..))))))))))..))))).))))..))). ( -32.32, z-score = -6.96, R) >triCas2.ChLG3 18613783 78 - 32080666 UUUUUGGUAGACCACUAGCGAGGUAUAGAGUUCCUACGUGAUUUUUCGGCU-----------GUAGGAACUUCAUACCGUGCUCUUGGA ...........(((..((((.(((((..(((((((((((((....)))..)-----------)))))))))..))))).))))..))). ( -31.90, z-score = -4.71, R) >anoGam1.chr2L 18352302 78 + 48795086 -----------CCACCAGCGAGGUAUAGAGUUCCUAUGUGCCCACUAGGCCGUUCGCUACUCAUAGGAACUUCAUACCGUGCUCUUGGA -----------(((..((((.(((((..((((((((((.(((.....))).((.....)).))))))))))..))))).))))..))). ( -33.10, z-score = -4.68, R) >apiMel3.Group7 4171611 67 + 9974240 -----------CCAGCAGCGAGGUAUAGAGUUCCUACGUAGUGUUCAGAAAGU-----------AGGAACUUCAUACCGUGCUCUUGGA -----------((((.((((.(((((..(((((((((..............))-----------)))))))..))))).)))).)))). ( -30.94, z-score = -5.95, R) >consensus _____UUUUUGCCAUCAGCGAGGUAUAGAGUUCCUACGUUCAUUUUAUAAA________CUCGUAGGAACUUCAUACCGUGCUCUUGGA ...........(((..((((.(((((..((((((((((.......................))))))))))..))))).))))..))). (-30.19 = -30.41 + 0.23)

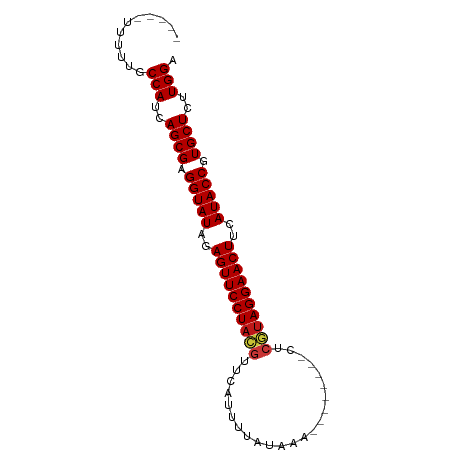
| Location | 10,358,332 – 10,358,406 |
|---|---|
| Length | 74 |
| Sequences | 15 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 85.48 |
| Shannon entropy | 0.31396 |
| G+C content | 0.42503 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.09 |
| Mean z-score | -5.42 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.998488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10358332 74 - 24543557 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAUAAUG--AACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------.........--..)))))))))).))))))..)))..)))......----- ( -28.02, z-score = -5.48, R) >droGri2.scaffold_15110 12043374 76 + 24565398 UCCAAGAGCACGGUAUGAAGUUCCUAUGAG--------UAUUUAAAUUUAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------((........)).)))))))))).))))))..)))..)))......----- ( -26.30, z-score = -5.11, R) >droMoj3.scaffold_6680 4142271 76 + 24764193 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UAUUUAAAUUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------((.......))..)))))))))).))))))..)))..)))......----- ( -29.00, z-score = -5.85, R) >droVir3.scaffold_13049 16143990 76 - 25233164 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UAUAUAAAUUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------((.......))..)))))))))).))))))..)))..)))......----- ( -29.00, z-score = -6.00, R) >droWil1.scaffold_181024 1528994 76 + 1751709 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAUUAAAUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.(((((((((..(--------(((((...))))))))))))))).))))))..)))..)))......----- ( -28.80, z-score = -5.77, R) >droPer1.super_27 1191224 76 - 1201981 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAUAAUAUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.(((((((((..(--------(((((...))))))))))))))).))))))..)))..)))......----- ( -28.80, z-score = -5.77, R) >dp4.chrXR_group6 8756412 76 - 13314419 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAUAAUAUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.(((((((((..(--------(((((...))))))))))))))).))))))..)))..)))......----- ( -28.80, z-score = -5.77, R) >droAna3.scaffold_13337 15274257 76 - 23293914 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAUAAUGUUAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------.(((......))))))))))))).))))))..)))..)))......----- ( -27.90, z-score = -5.21, R) >droEre2.scaffold_4784 10353140 74 - 25762168 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAUAAUG--AACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------.........--..)))))))))).))))))..)))..)))......----- ( -28.02, z-score = -5.48, R) >droYak2.chr3L 10348890 74 - 24197627 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAUA--AUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------......--.....)))))))))).))))))..)))..)))......----- ( -28.02, z-score = -5.48, R) >droSec1.super_0 2571170 74 - 21120651 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAUA--AUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------......--.....)))))))))).))))))..)))..)))......----- ( -28.02, z-score = -5.48, R) >droSim1.chr3L 9747999 74 - 22553184 UCCAAGAGCACGGUAUGAAGUUCCUACGAG--------UUUAU--AAUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA----- .(((..(((..((((((.((((((((((..--------.....--......)))))))))).))))))..)))..)))......----- ( -28.02, z-score = -5.48, R) >triCas2.ChLG3 18613783 78 + 32080666 UCCAAGAGCACGGUAUGAAGUUCCUAC-----------AGCCGAAAAAUCACGUAGGAACUCUAUACCUCGCUAGUGGUCUACCAAAAA .(((..(((..((((((.(((((((((-----------..............))))))))).))))))..)))..)))........... ( -27.74, z-score = -5.35, R) >anoGam1.chr2L 18352302 78 - 48795086 UCCAAGAGCACGGUAUGAAGUUCCUAUGAGUAGCGAACGGCCUAGUGGGCACAUAGGAACUCUAUACCUCGCUGGUGG----------- .(((..(((..((((((.((((((((((.((.....)).(((.....))).)))))))))).))))))..)))..)))----------- ( -33.70, z-score = -4.04, R) >apiMel3.Group7 4171611 67 - 9974240 UCCAAGAGCACGGUAUGAAGUUCCU-----------ACUUUCUGAACACUACGUAGGAACUCUAUACCUCGCUGCUGG----------- .(((..(((..((((((.(((((((-----------((..............))))))))).))))))..)))..)))----------- ( -25.54, z-score = -5.06, R) >consensus UCCAAGAGCACGGUAUGAAGUUCCUACGAG________UUUAUAAAAUGAACGUAGGAACUCUAUACCUCGCUGAUGGCAAAAA_____ .(((..(((..((((((.((((((((((.......................)))))))))).))))))..)))..)))........... (-26.54 = -26.64 + 0.10)
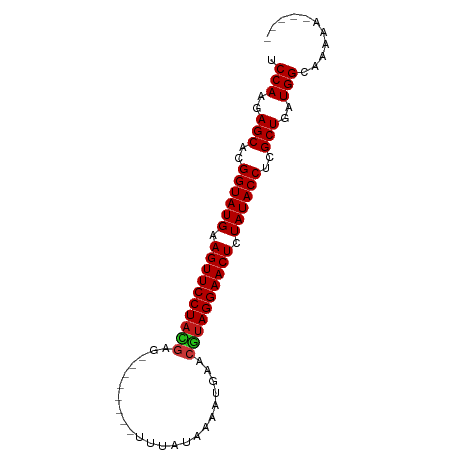
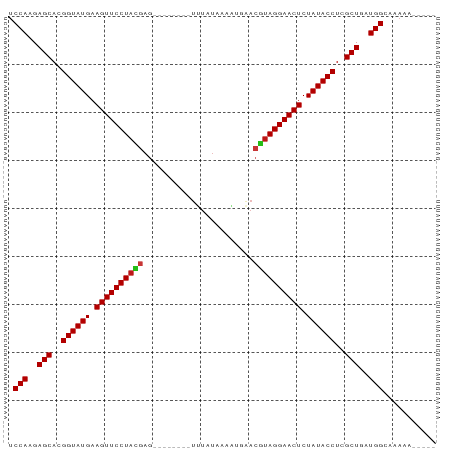
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:59 2011