| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,335,217 – 10,335,280 |
| Length | 63 |
| Max. P | 0.951774 |

| Location | 10,335,217 – 10,335,280 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 53.23 |
| Shannon entropy | 0.92625 |
| G+C content | 0.39813 |
| Mean single sequence MFE | -11.76 |
| Consensus MFE | -5.11 |
| Energy contribution | -4.74 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.75 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
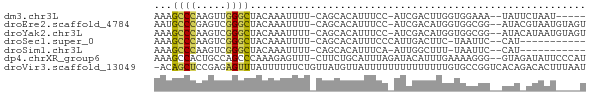
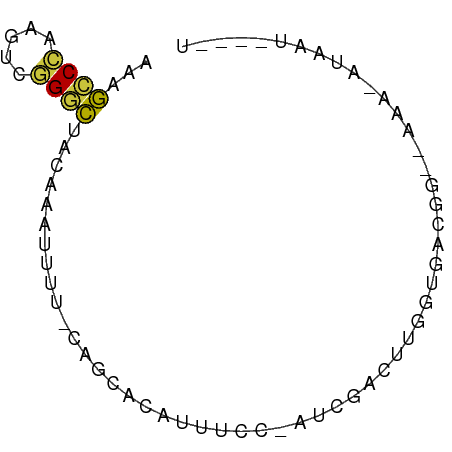
>dm3.chr3L 10335217 63 + 24543557 AAAGCCCAAGUUGGGCUACAAAUUUU-CAGCACAUUUCC-AUCGACUUGGUGGAAA--UAUUCUAAU----- ..(((((.....))))).........-......((((((-(((.....))))))))--)........----- ( -18.40, z-score = -3.46, R) >droEre2.scaffold_4784 10329097 68 + 25762168 AAUGCCCGAGUCGGGCUACAAAUUUU-CAGCACAUUUCC-AUCGACAUGGUGGCGG--AUACGUAAUGUAGU ...(((((...)))))..........-..(((((((..(-(((.....))))(((.--...)))))))).)) ( -14.40, z-score = 0.29, R) >droYak2.chr3L 10324696 68 + 24197627 AAAGCCCAAGUCGGGCUACAAAUUUU-CAGCACAUUUCC-AUCGACAUGGUGGCGG--AUACAUAAUGUAGU ..(((((.....))))).........-..(((((((.((-(((.....)))))...--......))))).)) ( -13.70, z-score = -0.44, R) >droSec1.super_0 2548615 57 + 21120651 AAAGCCCAAGUCGGGCUACAAAUUUU-CAGCACAUUUCCCAUUGACUUC-UAAUUC--CAU----------- ..(((((.....))))).........-......................-......--...----------- ( -8.50, z-score = -1.58, R) >droSim1.chr3L 9723725 56 + 22553184 AAAGCCCAAGUCGGGCUACAAAUUUU-CAGCACAUUUCA-AUUGGCUUU-UAAUUC--CAU----------- ..(((((.....)))))...((((..-.(((.((.....-..)))))..-.)))).--...----------- ( -10.90, z-score = -2.08, R) >dp4.chrXR_group6 8729990 69 + 13314419 AAAGCCACUGCCAGCCCAAAGAGUUU-CUUCUGCAUUUAGAUACAUUUGAAAAGGG--GUAGAUAUUCCCAU .......(((((....((((..((.(-((.........))).)).))))......)--)))).......... ( -9.80, z-score = 0.86, R) >droVir3.scaffold_13049 16115804 71 + 25233164 -ACAGCUCCGAGAGUUUAUUUUUUCUGUUAUGUUAUUUUUUUUUUUUUUGUGCCGGUCACAGACACUUUAAU -(((...(.(((((......))))).)...))).............((((((.....))))))......... ( -6.60, z-score = 0.22, R) >consensus AAAGCCCAAGUCGGGCUACAAAUUUU_CAGCACAUUUCC_AUCGACUUGGUGACGG__AAA_AUAAU____U ...((((.....))))........................................................ ( -5.11 = -4.74 + -0.36)
| Location | 10,335,217 – 10,335,280 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 53.23 |
| Shannon entropy | 0.92625 |
| G+C content | 0.39813 |
| Mean single sequence MFE | -12.42 |
| Consensus MFE | -4.53 |
| Energy contribution | -4.29 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10335217 63 - 24543557 -----AUUAGAAUA--UUUCCACCAAGUCGAU-GGAAAUGUGCUG-AAAAUUUGUAGCCCAACUUGGGCUUU -----.((((.(((--((((((.(.....).)-)))))))).)))-)........(((((.....))))).. ( -19.90, z-score = -3.66, R) >droEre2.scaffold_4784 10329097 68 - 25762168 ACUACAUUACGUAU--CCGCCACCAUGUCGAU-GGAAAUGUGCUG-AAAAUUUGUAGCCCGACUCGGGCAUU ....((.(((((.(--(((.(........).)-))).))))).))-..........(((((...)))))... ( -16.80, z-score = -1.08, R) >droYak2.chr3L 10324696 68 - 24197627 ACUACAUUAUGUAU--CCGCCACCAUGUCGAU-GGAAAUGUGCUG-AAAAUUUGUAGCCCGACUUGGGCUUU ..((((..((...(--(.((((((((....))-))...)).)).)-)..)).))))((((.....))))... ( -13.60, z-score = -0.29, R) >droSec1.super_0 2548615 57 - 21120651 -----------AUG--GAAUUA-GAAGUCAAUGGGAAAUGUGCUG-AAAAUUUGUAGCCCGACUUGGGCUUU -----------...--(((((.-..(((((........)).))).-..)))))..(((((.....))))).. ( -9.00, z-score = 0.40, R) >droSim1.chr3L 9723725 56 - 22553184 -----------AUG--GAAUUA-AAAGCCAAU-UGAAAUGUGCUG-AAAAUUUGUAGCCCGACUUGGGCUUU -----------...--(((((.-..(((((..-.....)).))).-..)))))..(((((.....))))).. ( -11.40, z-score = -1.13, R) >dp4.chrXR_group6 8729990 69 - 13314419 AUGGGAAUAUCUAC--CCCUUUUCAAAUGUAUCUAAAUGCAGAAG-AAACUCUUUGGGCUGGCAGUGGCUUU ..(((........)--))..((((...(((((....)))))...)-)))......(((((......))))). ( -10.20, z-score = 1.64, R) >droVir3.scaffold_13049 16115804 71 - 25233164 AUUAAAGUGUCUGUGACCGGCACAAAAAAAAAAAAAAUAACAUAACAGAAAAAAUAAACUCUCGGAGCUGU- ......(((((.......))))).....................((((...................))))- ( -6.01, z-score = 0.08, R) >consensus A____AUUAU_UAU__CCGCCACCAAGUCAAU_GGAAAUGUGCUG_AAAAUUUGUAGCCCGACUUGGGCUUU ........................................................(((((...)))))... ( -4.53 = -4.29 + -0.24)

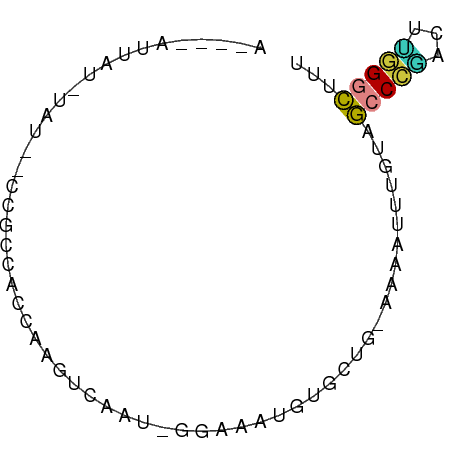
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:55 2011