| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,317,533 – 10,317,642 |
| Length | 109 |
| Max. P | 0.640146 |

| Location | 10,317,533 – 10,317,642 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 56.52 |
| Shannon entropy | 0.82789 |
| G+C content | 0.38434 |
| Mean single sequence MFE | -24.59 |
| Consensus MFE | -7.69 |
| Energy contribution | -7.28 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.94 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
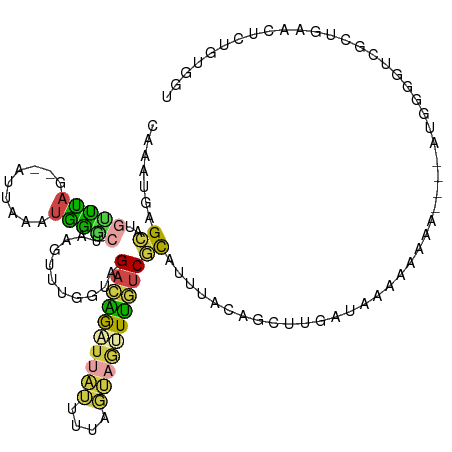
>dm3.chr3L 10317533 109 + 24543557 CAAAUGAGCAUGUUUAG--AUUAAAUGAGCUAAGUUUGGUAGACAGAUUAUUUUAGUAGUUUGUCGCCCUUACGAGUUGAUAAAAAAA-----AUGGGGUCGCCGAACUCUGUGGU ......(((.((((((.--..)))))).))).((((((((.((((((((((....))))))))))(((((......((....))....-----..))))).))))))))....... ( -28.20, z-score = -2.26, R) >droSec1.super_0 2531350 107 + 21120651 CAAAUGAGCAUGUUUAG--AUUAAAUGGGCUAAGUUUGGUAGACAGAUUAUUUUAGUAGUUUGUCGCCCUUGCGAGUUGAUAAAAA-------AUGGGGUCGCUGAACUCUGUGGU ......(((..(((((.--..)))))..))).((((..((.((((((((((....))))))))))(((((................-------..))))).))..))))....... ( -25.07, z-score = -0.83, R) >droYak2.chr3L 10307013 110 + 24197627 CAAAUGAGCAAGCUUAG--AUUAAAUGGGCUAAGUUUGGCGGACAGAUUAUUUUAGUAGUUUGUCGCAUUUACAGCUUGAUAAAAAAAA----ACCGGGUUGUUGAACUCUGUGGA ((...((((((((((((--..........)))))))))(((.(((((((((....)))))))))))).((.((((((((..........----..)))))))).)).)))..)).. ( -29.80, z-score = -2.43, R) >droEre2.scaffold_4784 10310938 114 + 25762168 CAUAUGAGCAAGCUUAG--AUUAAAUGGGCUAGGUUUGGCAGACAGUUUAUUUUAGUAGUUUGUCGCCUUUACGGCUUGAUAAAAAAAGUGGUAUGGGGUUGUUGAACUCUGUGGU ((((.((...((((((.--......))))))..(((..((((((((.((((....)))).))))).(((.(((.((((........)))).))).)))..)))..))))))))).. ( -31.60, z-score = -2.09, R) >droWil1.scaffold_181024 1467201 110 - 1751709 UCAAAAAGUCUGUUUAGGAAUUCAAUAGA--AAGAAAGAGUGAAGAGAAAGUUCAACACAUUUACAUAUUCAAAGAUGUAAACACUAAA----CUGAAACAACUAGAUCUAAUUCU ........((.((((((............--........(((..(((....)))..))).(((((((........)))))))..)))))----).))................... ( -14.30, z-score = 0.42, R) >droGri2.scaffold_15110 11997219 84 - 24565398 CCAAUGUGCAUGACCAA-----AAAAGGCGAAACUAGGCUAGAACGGUCGGCCCAUCCACUGGUCUUAUGAGCAGCUGUACAAGAUGAA--------------------------- ....(((((((((((..-----....(((........))).....))))).........(((.((....)).))).)))))).......--------------------------- ( -18.60, z-score = 0.23, R) >consensus CAAAUGAGCAUGUUUAG__AUUAAAUGGGCUAAGUUUGGUAGACAGAUUAUUUUAGUAGUUUGUCGCAUUUACAGCUUGAUAAAAAAAA____AUGGGGUCGCUGAACUCUGUGGU .......((..(((((.........)))))...........((((((((((....))))))))))))................................................. ( -7.69 = -7.28 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:54 2011