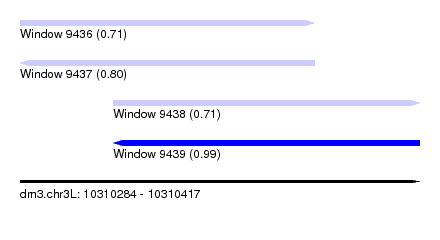
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,310,284 – 10,310,417 |
| Length | 133 |
| Max. P | 0.986492 |

| Location | 10,310,284 – 10,310,382 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.15 |
| Shannon entropy | 0.27998 |
| G+C content | 0.45065 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -12.35 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
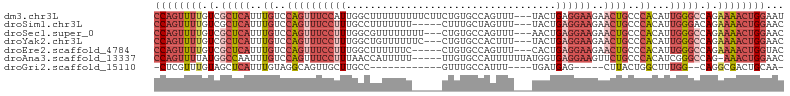
>dm3.chr3L 10310284 98 + 24543557 AUCCAUCAUCCCAUCCAGUUUUUCAGU--------GCCAAUUCCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUA---AAACUGGCACAGAAGAAAAAAAAAG ..................((((((.((--------((((....(((....)))((((.....))))...............---....)))))).))))))........ ( -24.00, z-score = -2.78, R) >droSim1.chr3L 9700329 93 + 22553184 AUCCAUCAUCCCAUCCAGUUUUUCAGU--------GCCAGUUCCAGUUUUCUGUCCCAAUGUGGGCAGUUCUUCCUCAGUA---AAACUAGCAAAGAAAAAAAG----- ..................((((((..(--------((.((((........((((((......)))))).............---.)))).)))..))))))...----- ( -15.85, z-score = -2.61, R) >droSec1.super_0 2524158 95 + 21120651 AUCCAUCAUCCCAUCCAGUUUUACAGU--------GCCAGUUCCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUU---AAACUGGCACAGAAAAAAAACG--- .................(((((...((--------(((((((.(((....)))((((.....))))...............---.)))))))))......))))).--- ( -25.40, z-score = -3.47, R) >droYak2.chr3L 10299511 103 + 24197627 AUCCAUCAUCCCGUCCAGUUUUUCAGUUUUUCAGUGCCAGUUCCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUA---AAAGUGGCACAGGAAAAAACAG--- ..........................((((((.((((((.((.(((....)))((((.....))))...............---.)).)))))).)))))).....--- ( -24.20, z-score = -2.36, R) >droEre2.scaffold_4784 10303524 93 + 25762168 AUCCAUCAUCCCGUCCAGUUUUUCAGU--------GCCAGUACCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUG---AAACUGGCACAGGAAAAAAG----- ..................((((((.((--------((((((..(((....)))((((.....))))...............---..)))))))).))))))...----- ( -27.80, z-score = -2.94, R) >droAna3.scaffold_13337 15231051 95 + 23293914 AUCCAUCAUCCCGUCCAGUUUCAGAGU--------UCCAGUUCCAGUUU-CUGGCCCGAUGUGGGCAGAACUUCCUCACCAUAAAAAAUGGCACAAAAAAAUGG----- ..((((.................(((.--------...((((((((...-)))((((.....)))).)))))..))).((((.....)))).........))))----- ( -18.20, z-score = -0.17, R) >consensus AUCCAUCAUCCCAUCCAGUUUUUCAGU________GCCAGUUCCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUA___AAACUGGCACAGAAAAAAAG_____ ..............(((((((......................(((....)))((((.....))))..................))))))).................. (-12.35 = -13.02 + 0.67)

| Location | 10,310,284 – 10,310,382 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Shannon entropy | 0.27998 |
| G+C content | 0.45065 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10310284 98 - 24543557 CUUUUUUUUUCUUCUGUGCCAGUUU---UACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGAAUUGGC--------ACUGAAAAACUGGAUGGGAUGAUGGAU .....((((((....((((((((((---(((((((......))((((.....))))))).....))))))))))--------)).)))))).................. ( -26.30, z-score = -1.39, R) >droSim1.chr3L 9700329 93 - 22553184 -----CUUUUUUUCUUUGCUAGUUU---UACUGAGGAAGAACUGCCCACAUUGGGACAGAAAACUGGAACUGGC--------ACUGAAAAACUGGAUGGGAUGAUGGAU -----(..((((((..(((((((((---((((.....))..((((((.....))).))).....))))))))))--------)..))))))..)............... ( -25.30, z-score = -3.25, R) >droSec1.super_0 2524158 95 - 21120651 ---CGUUUUUUUUCUGUGCCAGUUU---AACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGAACUGGC--------ACUGUAAAACUGGAUGGGAUGAUGGAU ---(((((.(((((.((((((((((---...............((((.....))))(((....)))))))))))--------)).).))))..)))))........... ( -27.90, z-score = -2.00, R) >droYak2.chr3L 10299511 103 - 24197627 ---CUGUUUUUUCCUGUGCCACUUU---UACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGAACUGGCACUGAAAAACUGAAAAACUGGACGGGAUGAUGGAU ---(((((...((((((.(((.(((---(..((.((........))))..(..((((((..........))))).)..)........)))).))))))))).))))).. ( -25.00, z-score = -1.49, R) >droEre2.scaffold_4784 10303524 93 - 25762168 -----CUUUUUUCCUGUGCCAGUUU---CACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGUACUGGC--------ACUGAAAAACUGGACGGGAUGAUGGAU -----((.((.((((((.(((((((---((.((....((.((.((((.....))))(((....))))).))..)--------).)))..)))))))))))).)).)).. ( -29.40, z-score = -2.14, R) >droAna3.scaffold_13337 15231051 95 - 23293914 -----CCAUUUUUUUGUGCCAUUUUUUAUGGUGAGGAAGUUCUGCCCACAUCGGGCCAG-AAACUGGAACUGGA--------ACUCUGAAACUGGACGGGAUGAUGGAU -----(((((.(((((((((((.....)))))(((..(((((.((((.....))))(((-...))))))))...--------.))).........)))))).))))).. ( -28.90, z-score = -1.74, R) >consensus _____CUUUUUUUCUGUGCCAGUUU___UACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGAACUGGC________ACUGAAAAACUGGACGGGAUGAUGGAU ...........((((((.(((((((..................((((.....))))(((....)))......................)))))))))))))........ (-18.72 = -18.72 + 0.00)

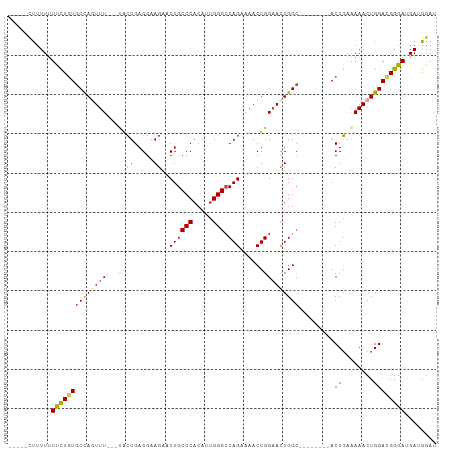
| Location | 10,310,315 – 10,310,417 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.75 |
| Shannon entropy | 0.46522 |
| G+C content | 0.45132 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -14.65 |
| Energy contribution | -16.83 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10310315 102 + 24543557 AUUCCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUA---AAACUGGCACAGAAGAAAAAAAAAGCCAAUGGAAACUGGACAAAUGAGCGACAAAACUGG ..((((((((((..((((.....))))..((((((.((((..---...))))....)))))).............)))))))))).................... ( -28.60, z-score = -1.83, R) >droSim1.chr3L 9700360 97 + 22553184 GUUCCAGUUUUCUGUCCCAAUGUGGGCAGUUCUUCCUCAGUA---AAACUAGCAAAG-----AAAAAAAGGCAAAGGAAACUGGACAAAUGAGCGACAAAACUGG ...((((((((((((((......))))))....((((((...---......((....-----........))..((....)).......)))).)).)))))))) ( -22.50, z-score = -0.77, R) >droSec1.super_0 2524189 99 + 21120651 GUUCCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUU---AAACUGGCACAG---AAAAAAAACGCCAAAGGAAACUGGACAAAUGAGCGACAAAACUGG ...((((((((...((((.....)))).((((.(((......---....((((....---.........)))).((....))))).....))))...)))))))) ( -27.72, z-score = -1.48, R) >droYak2.chr3L 10299550 99 + 24197627 GUUCCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUA---AAAGUGGCACAG---GAAAAAACAGCCAAAGGAAACUGGACAAAUGAGCGACAAAACUGG ...((((((((...((((.....)))).((((.(((......---....((((....---.........)))).((....))))).....))))...)))))))) ( -27.82, z-score = -1.06, R) >droEre2.scaffold_4784 10303555 97 + 25762168 GUACCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUG---AAACUGGCACAG-----GAAAAAAGCCAAAGGAAACUGGACAAAUGAGCGACAAAACUGG ...((((((((...((((.....)))).((((((((((((..---...)))....))-----)))....((((..(....)))).)....))))...)))))))) ( -29.00, z-score = -1.48, R) >droAna3.scaffold_13337 15231082 99 + 23293914 GUUCCAGUUU-CUGGCCCGAUGUGGGCAGAACUUCCUCACCAUAAAAAAUGGCACAA-----AAAAAUGGUUAAAGGAAACUGGACAAAUUGGCCAUAAAACUGG ...(((((((-.(((((.(((.((..(((...(((((.(((((..............-----....)))))...))))).)))..)).))))))))..))))))) ( -28.47, z-score = -2.15, R) >droGri2.scaffold_15110 11990408 80 - 24565398 -UUGCAGUCGCCUG--CCAAAGCCAGUAAG-----CUCAUCA----AAAUGGCAAAC------------GGCAAGCAACUGCCUACAAAUGAGCUACAAACGAG- -..(((((.((.((--((...((((.....-----.......----...))))....------------)))).)).)))))......................- ( -20.86, z-score = -1.42, R) >consensus GUUCCAGUUUUCUGGCCCAAUGUGGGCAGUUCUUCCUCAGUA___AAACUGGCACAG_____AAAAAAAGCCAAAGGAAACUGGACAAAUGAGCGACAAAACUGG ..(((((((((((.((((.....))))......................((((................)))).))))))))))).................... (-14.65 = -16.83 + 2.19)
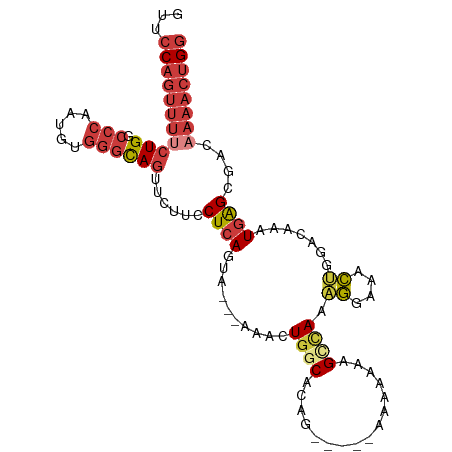
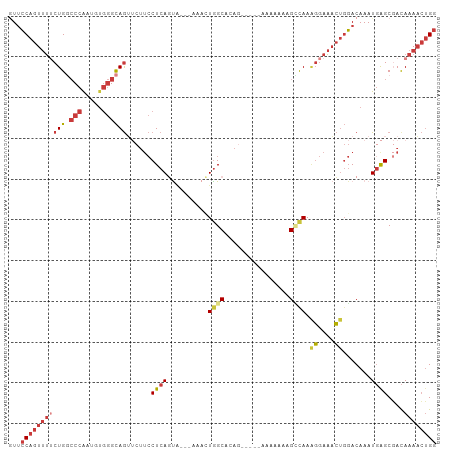
| Location | 10,310,315 – 10,310,417 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.75 |
| Shannon entropy | 0.46522 |
| G+C content | 0.45132 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -16.27 |
| Energy contribution | -18.30 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10310315 102 - 24543557 CCAGUUUUGUCGCUCAUUUGUCCAGUUUCCAUUGGCUUUUUUUUUCUUCUGUGCCAGUUU---UACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGAAU ((((((((.(.(((((..((..((((((((((((((................))))))..---......)))..)))))..))...))))).).))))))))... ( -30.79, z-score = -2.28, R) >droSim1.chr3L 9700360 97 - 22553184 CCAGUUUUGUCGCUCAUUUGUCCAGUUUCCUUUGCCUUUUUUU-----CUUUGCUAGUUU---UACUGAGGAAGAACUGCCCACAUUGGGACAGAAAACUGGAAC ....................((((((((.............((-----((((..(((...---..)))..))))))((((((.....))).))).)))))))).. ( -23.10, z-score = -0.77, R) >droSec1.super_0 2524189 99 - 21120651 CCAGUUUUGUCGCUCAUUUGUCCAGUUUCCUUUGGCGUUUUUUUU---CUGUGCCAGUUU---AACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGAAC ((((((((.(.(((((..((..(((((((((((((((........---...)))))....---....)))))..)))))..))...))))).).))))))))... ( -31.90, z-score = -2.52, R) >droYak2.chr3L 10299550 99 - 24197627 CCAGUUUUGUCGCUCAUUUGUCCAGUUUCCUUUGGCUGUUUUUUC---CUGUGCCACUUU---UACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGAAC ((((((((.(.(((((..((..((((((((((((((.........---....))))....---....)))))..)))))..))...))))).).))))))))... ( -31.92, z-score = -2.66, R) >droEre2.scaffold_4784 10303555 97 - 25762168 CCAGUUUUGUCGCUCAUUUGUCCAGUUUCCUUUGGCUUUUUUC-----CUGUGCCAGUUU---CACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGUAC ((((((((.(.(((((..((..((((((((((((((.......-----....))))....---....)))))..)))))..))...))))).).))))))))... ( -31.60, z-score = -2.53, R) >droAna3.scaffold_13337 15231082 99 - 23293914 CCAGUUUUAUGGCCAAUUUGUCCAGUUUCCUUUAACCAUUUUU-----UUGUGCCAUUUUUUAUGGUGAGGAAGUUCUGCCCACAUCGGGCCAG-AAACUGGAAC ((((((((.(((((....((..((((((((((..(((((....-----..............))))))))))))..)))..)).....))))))-)))))))... ( -32.47, z-score = -3.47, R) >droGri2.scaffold_15110 11990408 80 + 24565398 -CUCGUUUGUAGCUCAUUUGUAGGCAGUUGCUUGCC------------GUUUGCCAUUU----UGAUGAG-----CUUACUGGCUUUGG--CAGGCGACUGCAA- -((((..((.....))..)).))(((((((((((((------------(...((((.(.----.(.....-----)..).))))..)))--)))))))))))..- ( -32.20, z-score = -3.13, R) >consensus CCAGUUUUGUCGCUCAUUUGUCCAGUUUCCUUUGGCUUUUUUU_____CUGUGCCAGUUU___UACUGAGGAAGAACUGCCCACAUUGGGCCAGAAAACUGGAAC ((((((((.(.(((((..((..(((((((((.......................((((......)))))))))..))))..))...))))).).))))))))... (-16.27 = -18.30 + 2.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:50 2011