| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,287,410 – 10,287,500 |
| Length | 90 |
| Max. P | 0.992946 |

| Location | 10,287,410 – 10,287,500 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.43676 |
| G+C content | 0.41075 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -14.52 |
| Energy contribution | -15.04 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
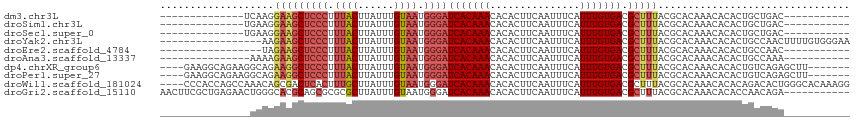
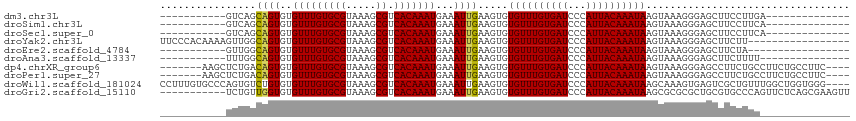
>dm3.chr3L 10287410 90 + 24543557 --------------UCAAGGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGCUGAC----------- --------------(((..(((((((((.((((......)))).))))(((((((...............))))))).)))))..(((.........)))))).----------- ( -19.46, z-score = -3.08, R) >droSim1.chr3L 9677025 90 + 22553184 --------------UGAAGGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGCUGAC----------- --------------((...(((((((((.((((......)))).))))(((((((...............))))))).)))))...))................----------- ( -19.36, z-score = -2.74, R) >droSec1.super_0 2501502 90 + 21120651 --------------UGAAGGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGCUGAC----------- --------------((...(((((((((.((((......)))).))))(((((((...............))))))).)))))...))................----------- ( -19.36, z-score = -2.74, R) >droYak2.chr3L 10275807 98 + 24197627 -----------------AAGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGCCAACUUUUGUGGGAA -----------------..(((((((((.((((......)))).))))(((((((...............))))))).))))).(.((((((.............)))))).).. ( -23.18, z-score = -3.03, R) >droEre2.scaffold_4784 10280544 87 + 25762168 -----------------UAGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGCCAAC----------- -----------------..(((((((((.((((......)))).))))(((((((...............))))))).)))))..(((.........)))....----------- ( -18.66, z-score = -3.80, R) >droAna3.scaffold_13337 15209446 89 + 23293914 ---------------AAAAGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGCCAAA----------- ---------------....(((((((((.((((......)))).))))(((((((...............))))))).)))))..(((.........)))....----------- ( -18.66, z-score = -3.38, R) >dp4.chrXR_group6 8681119 104 + 13314419 ----GAAGGCAGAAGGCAGAAGGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGUCAGAGCUU------- ----...(((((...((..(((((((((.((((......)))).))))(((((((...............))))))).)))))..)).........))))).......------- ( -24.66, z-score = -1.90, R) >droPer1.super_27 1121105 104 + 1201981 ----GAAGGCAGAAGGCAGAAGGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGUCAGAGCUU------- ----...(((((...((..(((((((((.((((......)))).))))(((((((...............))))))).)))))..)).........))))).......------- ( -24.66, z-score = -1.90, R) >droWil1.scaffold_181024 1427235 111 - 1751709 ----CCCACCAGCCAAACAGCGACUCACUUUGCUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACAGACACUGGGCACAAAGG ----.......(((....((((.((((..((((......)))))))).(((((((...............)))))))))))...............(((...))))))....... ( -19.36, z-score = -0.79, R) >droGri2.scaffold_15110 11966677 104 - 24565398 AACUUCGCUGAGAACUGGGCACGCAGCGCGCGCUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACCAACAGA----------- .......(((.....(((((.(((...))).)))))...((((.(.(.(((((((...............)))))))).).))))...............))).----------- ( -19.06, z-score = 0.30, R) >consensus _______________GAAGGAAGCUCCCUUUACUUAUUUGUAAUGGGAUCACAAACACACUUCAAUUUCAUUUGUGACGCUUUACGCACAAACACACUGCCAAC___________ ...................(((((((((.((((......)))).))))(((((((...............))))))).)))))................................ (-14.52 = -15.04 + 0.52)
| Location | 10,287,410 – 10,287,500 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.43676 |
| G+C content | 0.41075 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -16.72 |
| Energy contribution | -16.65 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
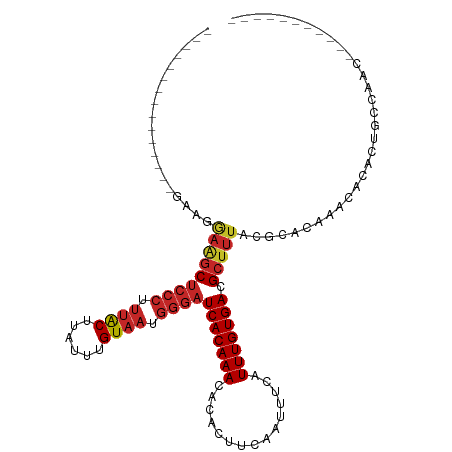
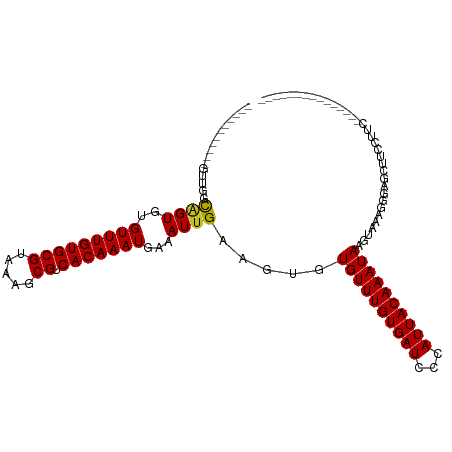
>dm3.chr3L 10287410 90 - 24543557 -----------GUCAGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCCUUGA-------------- -----------(..(((.....(((((((((.....)).))))))).....................((((.((((......)))).)))))))..)....-------------- ( -20.70, z-score = -1.28, R) >droSim1.chr3L 9677025 90 - 22553184 -----------GUCAGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCCUUCA-------------- -----------(..(((.....(((((((((.....)).))))))).....................((((.((((......)))).)))))))..)....-------------- ( -20.70, z-score = -1.35, R) >droSec1.super_0 2501502 90 - 21120651 -----------GUCAGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCCUUCA-------------- -----------(..(((.....(((((((((.....)).))))))).....................((((.((((......)))).)))))))..)....-------------- ( -20.70, z-score = -1.35, R) >droYak2.chr3L 10275807 98 - 24197627 UUCCCACAAAAGUUGGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCUU----------------- ....((((((......((((..(((((((((.....)).)))))))...)))).......)))))).((((.((((......)))).)))).......----------------- ( -26.52, z-score = -2.64, R) >droEre2.scaffold_4784 10280544 87 - 25762168 -----------GUUGGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCUA----------------- -----------...(((.....(((((((((.....)).))))))).....................((((.((((......)))).)))))))....----------------- ( -20.20, z-score = -1.22, R) >droAna3.scaffold_13337 15209446 89 - 23293914 -----------UUUGGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCUUUU--------------- -----------...(((.....(((((((((.....)).))))))).....................((((.((((......)))).)))))))......--------------- ( -20.20, z-score = -1.12, R) >dp4.chrXR_group6 8681119 104 - 13314419 -------AAGCUCUGACAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCCUUCUGCCUUCUGCCUUC---- -------..(((((..((((..(((((((((.....)).)))))))...)))).....((((((((((...)))))))))).......)))))..................---- ( -24.50, z-score = -1.23, R) >droPer1.super_27 1121105 104 - 1201981 -------AAGCUCUGACAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCCUUCUGCCUUCUGCCUUC---- -------..(((((..((((..(((((((((.....)).)))))))...)))).....((((((((((...)))))))))).......)))))..................---- ( -24.50, z-score = -1.23, R) >droWil1.scaffold_181024 1427235 111 + 1751709 CCUUUGUGCCCAGUGUCUGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGCAAAGUGAGUCGCUGUUUGGCUGGUGGG---- (((....((((((((.((....(((((((((.....)).))))))).........((.((((((((((...)))))))))).))......)).)))))...)))....)))---- ( -31.20, z-score = -1.98, R) >droGri2.scaffold_15110 11966677 104 + 24565398 -----------UCUGUUGGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGCGCGCGCUGCGUGCCCAGUUCUCAGCGAAGUU -----------..((((((...(((((((((.....)).)))))))......(((.((((((((((((...)))))))))).((((((...)))))).)).)))))))))..... ( -30.60, z-score = -1.65, R) >consensus ___________GUUGGCAGUGUGUUUGUGCGUAAAGCGUCACAAAUGAAAUUGAAGUGUGUUUGUGAUCCCAUUACAAAUAAGUAAAGGGAGCUUCCUUC_______________ ................((((..(((((((((.....)).)))))))...)))).....((((((((((...)))))))))).................................. (-16.72 = -16.65 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:43 2011