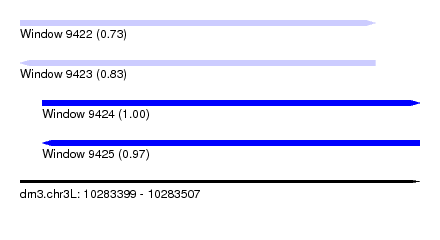
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,283,399 – 10,283,507 |
| Length | 108 |
| Max. P | 0.995187 |

| Location | 10,283,399 – 10,283,495 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Shannon entropy | 0.46677 |
| G+C content | 0.42657 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.733078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
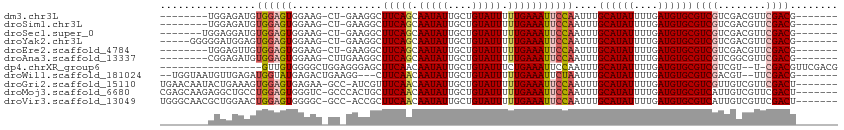
>dm3.chr3L 10283399 96 + 24543557 --------UGGAGAUGUGGAGUGGAAG-CU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGACGUUCGACG------- --------....(((.((((((.((((-(.-....)))))((((....))))...........))))))....((((((....)))))))))((((.....)))).------- ( -28.40, z-score = -1.75, R) >droSim1.chr3L 9673013 96 + 22553184 --------UGGAGAUGUGGAGUGGAAG-CU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGACGUUCGACG------- --------....(((.((((((.((((-(.-....)))))((((....))))...........))))))....((((((....)))))))))((((.....)))).------- ( -28.40, z-score = -1.75, R) >droSec1.super_0 2497530 97 + 21120651 -------UGGAGGAUGUGGAGUGGAAG-CU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGACGUUCGACG------- -------.(..((((((.((.(((((.-.(-.((((...(((((....)))))...)))).)..)))))....((((((....))))))....)).))))))..).------- ( -28.70, z-score = -1.72, R) >droYak2.chr3L 10271802 99 + 24197627 -----GGGGGAUGGAGUGGAGUGGAAG-CU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGACGUUCGACG------- -----....(((....((((((.((((-(.-....)))))((((....))))...........))))))....((((((....)))))))))((((.....)))).------- ( -28.20, z-score = -1.26, R) >droEre2.scaffold_4784 10276455 96 + 25762168 --------UGGAGUUGUGGAGUGGAAG-CU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGACGUUCGACG------- --------(((((((..((((((((((-(.-....)))))((((....)))).))))))...)))))))....((((((....))))))..(((((.....)))))------- ( -30.90, z-score = -2.62, R) >droAna3.scaffold_13337 15205859 97 + 23293914 --------CGGAGAUGUGGAGUGGAAG-CUUGAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGGCGUUCGACG------- --------((((((((..((.(((((.-.(..((((...(((((....)))))..))))..)..))))).))..))).))))).....(((((........)))))------- ( -28.90, z-score = -1.53, R) >dp4.chrXR_group6 8676808 93 + 13314419 -----------------GUUGUGGGGCUGGAGGGAGCUUCAACAAUAUUGCUGUAUUUCUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGU--U-CGACGUUCGACG -----------------...((.((((.....((((.((((..(((((....)))))..)))).)))).....((((((....))))))((((....--.-)))))))).)). ( -25.40, z-score = -0.97, R) >droWil1.scaffold_181024 1423612 99 - 1751709 --UGGUAAUGUUGAGAUGGUAUGAGACUGAAGG---CUUCAACAAUAUUGCUGUAUUUUUGAAAUUCUAAUUUGCAUAUUUUGAUGUGCGUCGACGU--UUCGACG------- --.......(((((((((......(((.(((..---.(((((.(((((....))))).))))).)))......((((((....)))))))))..)))--)))))).------- ( -24.70, z-score = -1.68, R) >droGri2.scaffold_15110 11963220 104 - 24565398 UGAACAAUACUGAAAGUGGAGUGAGAA-GCC-AUCGUUUCAACAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUUGUCGUUCGACU------- .((((.(((.(((...((((((((...-...-.))..(((((.(((((....))))).)))))))))))....((((((....)))))).))).))).))))....------- ( -22.60, z-score = -0.36, R) >droMoj3.scaffold_6680 4053663 105 - 24764193 CGAGCAAGAGGCUGCCUGGAGUGGGUC-GCCCACUGCUUCAACAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCAUUGUCGUUCGACU------- (((((..(((((....(((((((((..-.)))))...(((((.(((((....))))).)))))..))))....((((((....))))))))).))...)))))...------- ( -28.80, z-score = -0.91, R) >droVir3.scaffold_13049 16062831 104 + 25233164 UGGGCAACGCUGGAACUGGAGUGGGGC-GCC-ACCGCUUCAACAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCAUUGUCGUUCGACU------- ..(((.....(((((.(((((((((..-..)-.))))))))..(((((....))))).......)))))....((((((....)))))))))...(((....))).------- ( -26.90, z-score = -0.06, R) >consensus ________UGGAGAUGUGGAGUGGAAG_CU_GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGACGUUCGACG_______ ................((((((...............(((((.(((((....))))).)))))))))))....((((((....))))))((((........))))........ (-16.20 = -16.48 + 0.27)
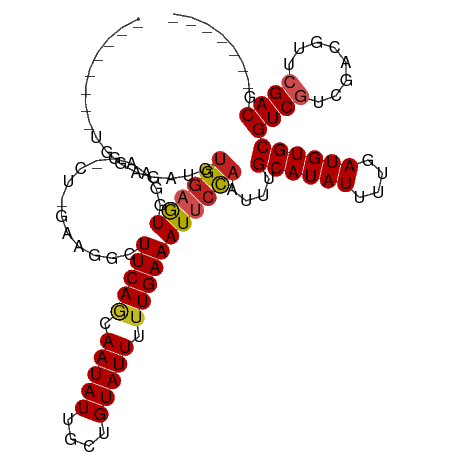
| Location | 10,283,399 – 10,283,495 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Shannon entropy | 0.46677 |
| G+C content | 0.42657 |
| Mean single sequence MFE | -19.26 |
| Consensus MFE | -10.85 |
| Energy contribution | -10.45 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10283399 96 - 24543557 -------CGUCGAACGUCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AG-CUUCCACUCCACAUCUCCA-------- -------(((((.....)))))..(((.((....)).)))....((((.............((((....))))(((((....-.)-))))...))))........-------- ( -20.10, z-score = -2.43, R) >droSim1.chr3L 9673013 96 - 22553184 -------CGUCGAACGUCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AG-CUUCCACUCCACAUCUCCA-------- -------(((((.....)))))..(((.((....)).)))....((((.............((((....))))(((((....-.)-))))...))))........-------- ( -20.10, z-score = -2.43, R) >droSec1.super_0 2497530 97 - 21120651 -------CGUCGAACGUCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AG-CUUCCACUCCACAUCCUCCA------- -------(((((.....)))))..(((.((....)).)))....((((.............((((....))))(((((....-.)-))))...)))).........------- ( -20.10, z-score = -2.53, R) >droYak2.chr3L 10271802 99 - 24197627 -------CGUCGAACGUCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AG-CUUCCACUCCACUCCAUCCCCC----- -------(((((.....)))))..(((.((....)).)))....((((.............((((....))))(((((....-.)-))))........))))......----- ( -20.20, z-score = -2.89, R) >droEre2.scaffold_4784 10276455 96 - 25762168 -------CGUCGAACGUCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AG-CUUCCACUCCACAACUCCA-------- -------(((((.....)))))..(((.((....)).)))....((((.............((((....))))(((((....-.)-))))...))))........-------- ( -20.10, z-score = -2.53, R) >droAna3.scaffold_13337 15205859 97 - 23293914 -------CGUCGAACGCCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUCAAG-CUUCCACUCCACAUCUCCG-------- -------(((((.....)))))..(((.((....)).)))....((((.............((((....))))(((((......)-))))...))))........-------- ( -19.70, z-score = -2.41, R) >dp4.chrXR_group6 8676808 93 - 13314419 CGUCGAACGUCG-A--ACGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAGAAAUACAGCAAUAUUGUUGAAGCUCCCUCCAGCCCCACAAC----------------- .((((..((((.-.--..)))).)).))..........((.....(((.((((((.((((......)))).)))))).))).....))........----------------- ( -14.80, z-score = -0.30, R) >droWil1.scaffold_181024 1423612 99 + 1751709 -------CGUCGAA--ACGUCGACGCACAUCAAAAUAUGCAAAUUAGAAUUUCAAAAAUACAGCAAUAUUGUUGAAG---CCUUCAGUCUCAUACCAUCUCAACAUUACCA-- -------.(((((.--...)))))(((.((....)).)))......(((((((((.((((......)))).))))))---..)))..........................-- ( -12.20, z-score = -0.66, R) >droGri2.scaffold_15110 11963220 104 + 24565398 -------AGUCGAACGACAACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGUUGAAACGAU-GGC-UUCUCACUCCACUUUCAGUAUUGUUCA -------....((((((....((.((.((((............((((....)))).....(((((....)))))....)))-)))-.))..(((........))).)))))). ( -17.30, z-score = -0.24, R) >droMoj3.scaffold_6680 4053663 105 + 24764193 -------AGUCGAACGACAAUGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGUUGAAGCAGUGGGC-GACCCACUCCAGGCAGCCUCUUGCUCG -------...(((.((((((((..(((.((....)).)))...((((....))))...........))))))))..((((.((((-..((.......))..)))).))))))) ( -23.00, z-score = -0.43, R) >droVir3.scaffold_13049 16062831 104 - 25233164 -------AGUCGAACGACAAUGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGUUGAAGCGGU-GGC-GCCCCACUCCAGUUCCAGCGUUGCCCA -------.(((....)))...(((((.(((......))).....(((((((.........(((((....)))))..(.(((-((.-...))))).)))))))))))))..... ( -24.30, z-score = -0.70, R) >consensus _______CGUCGAACGACGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC_AG_CUUCCACUCCACAUCUCCA________ ........((((........))))(((.((....)).)))....................(((((....)))))....................................... (-10.85 = -10.45 + -0.40)

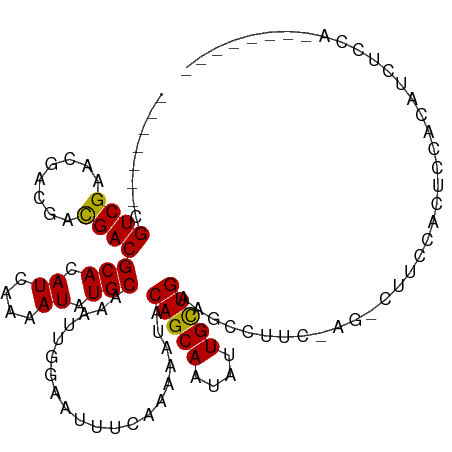
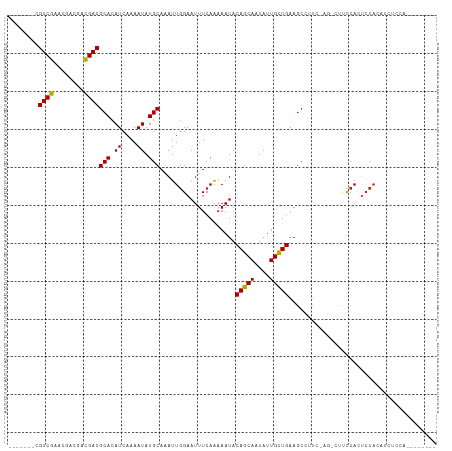
| Location | 10,283,405 – 10,283,507 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.03 |
| Shannon entropy | 0.37771 |
| G+C content | 0.43650 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.69 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10283405 102 + 24543557 ------UGUGGAGUGGAAGCU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGA----CGUUCGACGCUUCCACUCCCU- ------...(((((((((((.-...(((..(((((....)))))....................((((((....)))))))))((((.----....)))))))))))))))..- ( -40.90, z-score = -5.50, R) >droSim1.chr3L 9673019 102 + 22553184 ------UGUGGAGUGGAAGCU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGA----CGUUCGACGCUUCCACUCCCU- ------...(((((((((((.-...(((..(((((....)))))....................((((((....)))))))))((((.----....)))))))))))))))..- ( -40.90, z-score = -5.50, R) >droSec1.super_0 2497537 102 + 21120651 ------UGUGGAGUGGAAGCU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGA----CGUUCGACGCUUCCACUCCCU- ------...(((((((((((.-...(((..(((((....)))))....................((((((....)))))))))((((.----....)))))))))))))))..- ( -40.90, z-score = -5.50, R) >droYak2.chr3L 10271808 106 + 24197627 ---UGGAGUGGAGUGGAAGCU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGA----CGUUCGACGCUUCCACUCCCUU ---..(((.(((((((((((.-...(((..(((((....)))))....................((((((....)))))))))((((.----....)))))))))))))))))) ( -42.90, z-score = -5.33, R) >droEre2.scaffold_4784 10276461 102 + 25762168 ------UGUGGAGUGGAAGCU-GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGA----CGUUCGACGCUUCCACUCCCU- ------...(((((((((((.-...(((..(((((....)))))....................((((((....)))))))))((((.----....)))))))))))))))..- ( -40.90, z-score = -5.50, R) >droAna3.scaffold_13337 15205865 103 + 23293914 ------UGUGGAGUGGAAGCUUGAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGG----CGUUCGACGCUUCCACUUACU- ------....(((((((((((((((.(((.(((((....)))))....................((((((....))))))......))----).))))).))))))))))...- ( -37.20, z-score = -3.98, R) >dp4.chrXR_group6 8676810 103 + 13314419 ------UGUGGGGCUGGAGGG----AGCUUCAACAAUAUUGCUGUAUUUCUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGUUCGACGUUCGACGCUGCCACUCAGU- ------.(((((((.....((----((.((((..(((((....)))))..)))).)))).....((((((....))))))((((.((.....))..))))))).)))).....- ( -29.60, z-score = -0.76, R) >droWil1.scaffold_181024 1423618 104 - 1751709 UGUUGAGAUGGUAUGAGACUG--AAGGCUUCAACAAUAUUGCUGUAUUUUUGAAAUUCUAAUUUGCAUAUUUUGAUGUGCGUCGACGU------UUCGACGGUCUGACUCGA-- .(((.((((.((.((((((.(--..((((((((.(((((....))))).)))))..........((((((....)))))))))..)))------)))))).)))))))....-- ( -26.60, z-score = -1.39, R) >droGri2.scaffold_15110 11963230 106 - 24565398 --UGAAAGUGGAGUGAGAAGCC-AUCGUUUCAACAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUUGU----CGUUCGACUGAGCCACCAACG- --..(((.((((((((......-.))..(((((.(((((....))))).))))))))))).)))((((((....))))))..(((((.----.((((....))))...)))))- ( -22.70, z-score = 0.01, R) >droMoj3.scaffold_6680 4053675 105 - 24764193 ----UGCCUGGAGUGGGUCGCCCACUGCUUCAACAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCAUUGU----CGUUCGACUCUGCCACCAGCC- ----...((((((((((...))))))..(((((.(((((....))))).)))))..........((((((....))))))(.((..((----(....)))..)).).))))..- ( -26.60, z-score = -1.13, R) >droVir3.scaffold_13049 16062841 106 + 25233164 --UGGAACUGGAGUGGGGCGCC-ACCGCUUCAACAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCAUUGU----CGUUCGACUCUACCGCCAACC- --(((...((((((.(((((.(-(..(((((((.(((((....))))).)))))..........((((((....))))))))...)).----))))).))))))...)))...- ( -29.70, z-score = -1.65, R) >consensus ______UGUGGAGUGGAAGCU_GAAGGCUUCAGCAAUAUUGCUGUAUUUUUGAAAUUCCAAUUUGCAUAUUUUGAUGUGCGUCGUCGA____CGUUCGACGCUUCCACUCCCU_ ............((((.(((........(((((.(((((....))))).)))))..........((((((....))))))((((.((.....))..))))))).))))...... (-18.41 = -18.69 + 0.28)

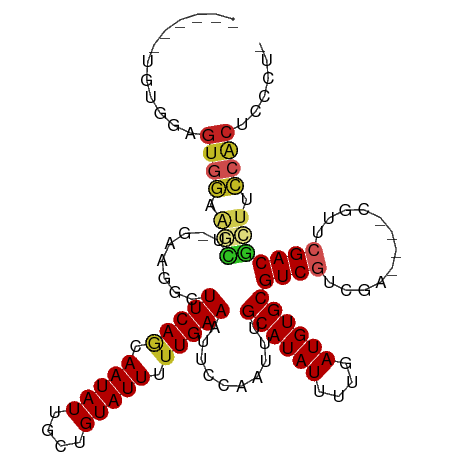
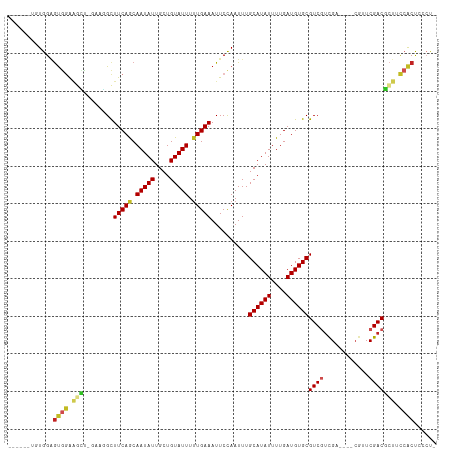
| Location | 10,283,405 – 10,283,507 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.03 |
| Shannon entropy | 0.37771 |
| G+C content | 0.43650 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -13.42 |
| Energy contribution | -12.95 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.37 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10283405 102 - 24543557 -AGGGAGUGGAAGCGUCGAACG----UCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AGCUUCCACUCCACA------ -..(((((((((((((((....----.))))((.((.................((((....)))).....(((((....)))))..))..))-.)))))))))))...------ ( -36.60, z-score = -5.25, R) >droSim1.chr3L 9673019 102 - 22553184 -AGGGAGUGGAAGCGUCGAACG----UCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AGCUUCCACUCCACA------ -..(((((((((((((((....----.))))((.((.................((((....)))).....(((((....)))))..))..))-.)))))))))))...------ ( -36.60, z-score = -5.25, R) >droSec1.super_0 2497537 102 - 21120651 -AGGGAGUGGAAGCGUCGAACG----UCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AGCUUCCACUCCACA------ -..(((((((((((((((....----.))))((.((.................((((....)))).....(((((....)))))..))..))-.)))))))))))...------ ( -36.60, z-score = -5.25, R) >droYak2.chr3L 10271808 106 - 24197627 AAGGGAGUGGAAGCGUCGAACG----UCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AGCUUCCACUCCACUCCA--- ...(((((((((((((((....----.))))((.((.................((((....)))).....(((((....)))))..))..))-.)))))))))))......--- ( -36.60, z-score = -4.96, R) >droEre2.scaffold_4784 10276461 102 - 25762168 -AGGGAGUGGAAGCGUCGAACG----UCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC-AGCUUCCACUCCACA------ -..(((((((((((((((....----.))))((.((.................((((....)))).....(((((....)))))..))..))-.)))))))))))...------ ( -36.60, z-score = -5.25, R) >droAna3.scaffold_13337 15205865 103 - 23293914 -AGUAAGUGGAAGCGUCGAACG----CCGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUCAAGCUUCCACUCCACA------ -.((.(((((((((((((....----.))))((.((.................((((....)))).....(((((....)))))..))..))..)))))))))..)).------ ( -29.50, z-score = -3.65, R) >dp4.chrXR_group6 8676810 103 - 13314419 -ACUGAGUGGCAGCGUCGAACGUCGAACGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAGAAAUACAGCAAUAUUGUUGAAGCU----CCCUCCAGCCCCACA------ -.....((((..((((((..((.....)).))))))............((.....(((.((((((.((((......)))).)))))).)----)).....)).)))).------ ( -25.20, z-score = -1.50, R) >droWil1.scaffold_181024 1423618 104 + 1751709 --UCGAGUCAGACCGUCGAA------ACGUCGACGCACAUCAAAAUAUGCAAAUUAGAAUUUCAAAAAUACAGCAAUAUUGUUGAAGCCUU--CAGUCUCAUACCAUCUCAACA --..(((.(.....(((((.------...)))))(((.((....)).)))......(((((((((.((((......)))).))))))..))--).).))).............. ( -16.30, z-score = -0.99, R) >droGri2.scaffold_15110 11963230 106 + 24565398 -CGUUGGUGGCUCAGUCGAACG----ACAACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGUUGAAACGAU-GGCUUCUCACUCCACUUUCA-- -.(..(((((....((((....----....))))((.((((............((((....)))).....(((((....)))))....)))-)))........)))))..).-- ( -22.50, z-score = -1.03, R) >droMoj3.scaffold_6680 4053675 105 + 24764193 -GGCUGGUGGCAGAGUCGAACG----ACAAUGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGUUGAAGCAGUGGGCGACCCACUCCAGGCA---- -(.((((((.((..(((....)----))..)).)))...........(((...((((....)))).....(((((....)))))..)))(((((...)))))..))).).---- ( -25.50, z-score = -0.47, R) >droVir3.scaffold_13049 16062841 106 - 25233164 -GGUUGGCGGUAGAGUCGAACG----ACAAUGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGUUGAAGCGGU-GGCGCCCCACUCCAGUUCCA-- -(((..(((.((..(((....)----))..)).)))..))).............(((((((.........(((((....)))))..(.(((-((....))))).))))))))-- ( -23.40, z-score = 0.44, R) >consensus _AGGGAGUGGAAGCGUCGAACG____ACGACGACGCACAUCAAAAUAUGCAAAUUGGAAUUUCAAAAAUACAGCAAUAUUGCUGAAGCCUUC_AGCUUCCACUCCACA______ .....(((((....((((............))))(((.((....)).)))....................(((((....)))))..............)))))........... (-13.42 = -12.95 + -0.48)

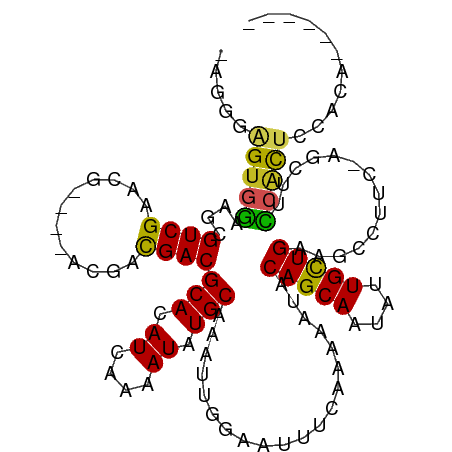
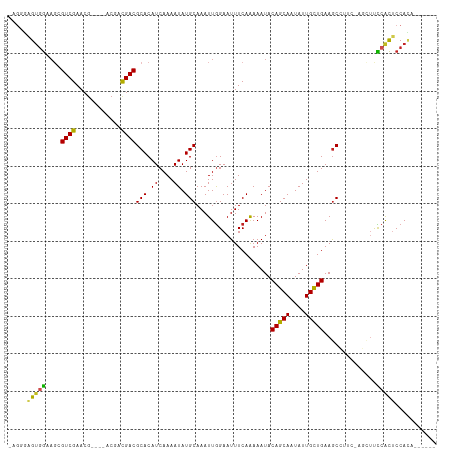
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:39 2011