| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,730,229 – 4,730,311 |
| Length | 82 |
| Max. P | 0.568640 |

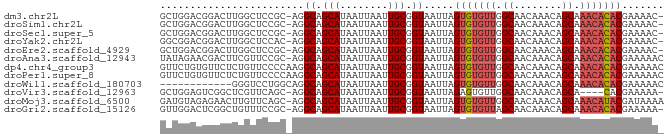
| Location | 4,730,229 – 4,730,311 |
|---|---|
| Length | 82 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 84.82 |
| Shannon entropy | 0.36013 |
| G+C content | 0.44394 |
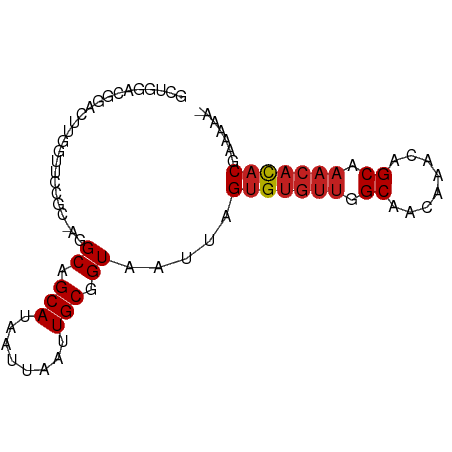
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.37 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
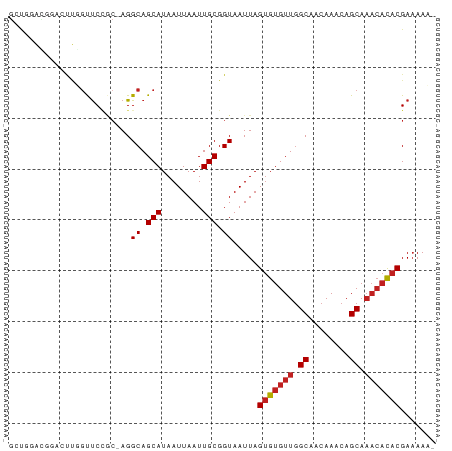
>dm3.chr2L 4730229 82 + 23011544 GCUGGACGGACUUGGCUCCGC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAC- (((.(.((((......)))))-.))).(((........)))........(((((((.((........)).)))))))......- ( -23.30, z-score = -1.74, R) >droSim1.chr2L 4598757 82 + 22036055 GCUGGACGGACUUGGCUCCGC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAC- (((.(.((((......)))))-.))).(((........)))........(((((((.((........)).)))))))......- ( -23.30, z-score = -1.74, R) >droSec1.super_5 2806759 82 + 5866729 GCUGGACGGACUUGGCUCCGC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAC- (((.(.((((......)))))-.))).(((........)))........(((((((.((........)).)))))))......- ( -23.30, z-score = -1.74, R) >droYak2.chr2L 4738064 82 + 22324452 GGCGGACGGACUUGGCUCCAC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAC- .((....(((......)))..-..)).(((........)))........(((((((.((........)).)))))))......- ( -18.00, z-score = -0.41, R) >droEre2.scaffold_4929 4805289 82 + 26641161 GCUGGACGGACUUGGCUCCGC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAC- (((.(.((((......)))))-.))).(((........)))........(((((((.((........)).)))))))......- ( -23.30, z-score = -1.74, R) >droAna3.scaffold_12943 1932148 83 - 5039921 UAUAGAACGACUUCGUUCCGC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAAC ....(((((....)))))(((-((.............))))).......(((((((.((........)).)))))))....... ( -21.42, z-score = -2.60, R) >dp4.chr4_group3 579317 84 - 11692001 GUUCUGUGUUCUCUGUUCCCCAAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAAC .(((((((((..(((((......(.(((((((.(((((((...)))))))))))))).)....)))))..)))))).))).... ( -22.20, z-score = -1.95, R) >droPer1.super_8 1623373 84 - 3966273 GUUCUGUGUUCUCUGUUCCCCAAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAAC .(((((((((..(((((......(.(((((((.(((((((...)))))))))))))).)....)))))..)))))).))).... ( -22.20, z-score = -1.95, R) >droWil1.scaffold_180703 1469566 72 - 3946847 ------------GGGUCCUGGCAGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAAC ------------..(((......))).(((........)))........(((((((.((........)).)))))))....... ( -16.00, z-score = -0.63, R) >droVir3.scaffold_12963 11553601 78 + 20206255 GCUGGAGUCGGCUCGUUCAGC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGAGUGUUGGCAACAAACAGCA----CACGAAAAA- (((((((....)))...))))-..((.(((........))).)).......((((((........)))))----)........- ( -19.10, z-score = -0.73, R) >droMoj3.scaffold_6500 20241222 83 + 32352404 GAUGUAGAGAACUUGUUCAGC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACAUACGAUAAAA ..(((.(((......))).))-).((.(((........))).)).....(((((((.((........)).)))))))....... ( -15.60, z-score = -0.16, R) >droGri2.scaffold_15126 5125325 82 + 8399593 GUUGGACUCGGCUGUUUCCGC-AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUAGCAACAAACAGCAAACACACGAAAAA- .....(((.(.((((....))-)).).(((........)))))).....(((((((.((........)).)))))))......- ( -20.20, z-score = -0.89, R) >consensus GCUGGACGGACUUGGUUCCGC_AGGCAGCAUAAUUAAUUGCGGUAAUUAGUGUGUUGGCAACAAACAGCAAACACACGAAAAA_ ........................((.(((........))).)).....(((((((.((........)).)))))))....... (-14.03 = -14.37 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:20 2011