| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,272,084 – 10,272,174 |
| Length | 90 |
| Max. P | 0.986833 |

| Location | 10,272,084 – 10,272,174 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.68 |
| Shannon entropy | 0.27588 |
| G+C content | 0.38727 |
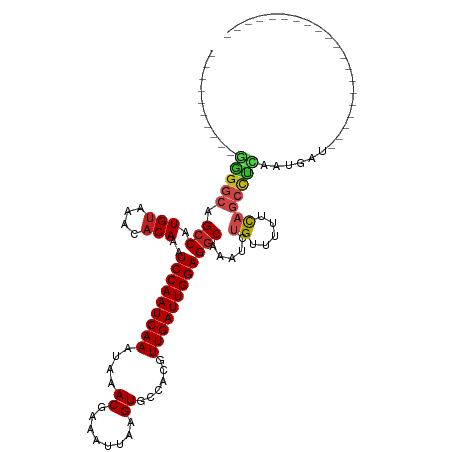
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -19.89 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10272084 90 - 24543557 ----------GGGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUCAGCCUCAAUGAU-------------------- ----------(((((.(((.(((....)))..(((((((((....((........))......)))))))))))).....((.....)))))))......-------------------- ( -25.10, z-score = -2.76, R) >droSim1.chr3L 9661618 90 - 22553184 ----------GGGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUCAGCCUCAAUGAU-------------------- ----------(((((.(((.(((....)))..(((((((((....((........))......)))))))))))).....((.....)))))))......-------------------- ( -25.10, z-score = -2.76, R) >droSec1.super_0 2486652 90 - 21120651 ----------GGGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUCAGCCUCAAUGAU-------------------- ----------(((((.(((.(((....)))..(((((((((....((........))......)))))))))))).....((.....)))))))......-------------------- ( -25.10, z-score = -2.76, R) >droYak2.chr3L 10260328 90 - 24197627 ----------GGGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUCAGCCUCAAUGAU-------------------- ----------(((((.(((.(((....)))..(((((((((....((........))......)))))))))))).....((.....)))))))......-------------------- ( -25.10, z-score = -2.76, R) >droEre2.scaffold_4784 10264224 90 - 25762168 ----------GGGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUCAGCCUCAAUGAU-------------------- ----------(((((.(((.(((....)))..(((((((((....((........))......)))))))))))).....((.....)))))))......-------------------- ( -25.10, z-score = -2.76, R) >droAna3.scaffold_13337 15191922 90 - 23293914 ----------GAGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUCAGCCUCAAUGAU-------------------- ----------(((((.(((.(((....)))..(((((((((....((........))......)))))))))))).....((.....)))))))......-------------------- ( -26.00, z-score = -3.67, R) >dp4.chrXR_group6 8664548 120 - 13314419 UUCCGGCUAGAGGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUUCUCUCUCUUUUUUUUUCUUCAGCCUCAAUGAU ....((((.(((((..(((.(((....)))..(((((((((....((........))......))))))))))))(((......)))................)))))))))........ ( -25.60, z-score = -1.01, R) >droPer1.super_27 1104627 119 - 1201981 UUCCGGCUAGAGGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUUCUCUCUCUUUUUUUUU-UUCAGCCUCAAUGAU ....((((((((((..(((.(((....)))..(((((((((....((........))......))))))))))))(((......)))...))))))........-...))))........ ( -25.20, z-score = -0.91, R) >droWil1.scaffold_181024 1407754 89 + 1751709 ----------GAGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUG-CACGUUGAUUGGAGGCAAAUCUGUUUUUCAGGCUGAAUGAU-------------------- ----------....(((((.((.(((((....(((((((((....((........)).-....)))))))))........)))))..)))))))......-------------------- ( -23.60, z-score = -2.51, R) >droVir3.scaffold_13049 16052563 89 - 25233164 ----------UAGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUG-CACGUUGAUUGGAGCCAAAUCUGUUUUUCAGCCUGAAUGAU-------------------- ----------(((((.....(((....)))..(((((((((....((........)).-....))))))))).................)))))......-------------------- ( -19.50, z-score = -1.75, R) >droMoj3.scaffold_6680 4042478 88 + 24764193 ----------UAGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUG-CACGUUGAUUGGAGGCAAAUCUG-UUUUCAGCCUGAAUGAU-------------------- ----------(((((.(((.(((....)))..(((((((((....((........)).-....)))))))))))).....((-....)))))))......-------------------- ( -23.80, z-score = -3.09, R) >droGri2.scaffold_15110 11952815 84 + 24565398 --------------CAGCCAUGAAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUG-CACGUUGAUUGGAGGCAAAUCUG-UUUUCAGCCUGAAUGAU-------------------- --------------(((...((((((((....(((((((((....((........)).-....)))))))))........))-))))))..)))......-------------------- ( -21.80, z-score = -3.13, R) >consensus __________GGGGCAGCCAUGUAAACACAAAUCCAAUCAAAUAAACGAAAUUAAGUGCCACGUUGAUUGGAGGCAAAUCUGUUUUUCAGCCUCAAUGAU____________________ ..........(((((.(((.(((....)))..(((((((((....((........))......)))))))))))).....((.....))))))).......................... (-19.89 = -19.92 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:33 2011