| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,248,906 – 10,249,023 |
| Length | 117 |
| Max. P | 0.770144 |

| Location | 10,248,906 – 10,249,023 |
|---|---|
| Length | 117 |
| Sequences | 11 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Shannon entropy | 0.49816 |
| G+C content | 0.35426 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.26 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
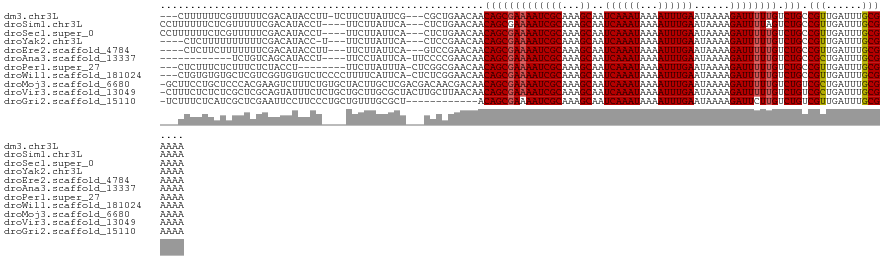
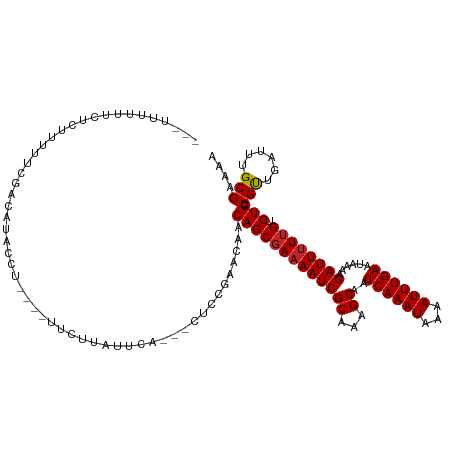
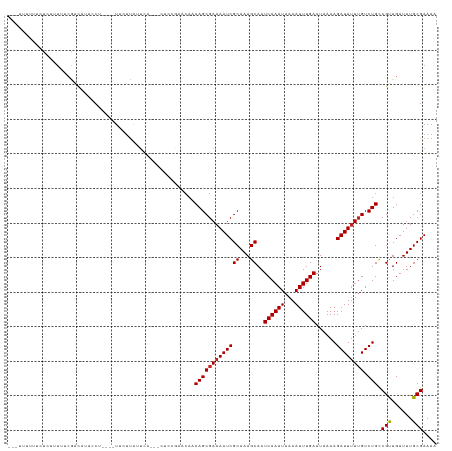
>dm3.chr3L 10248906 117 - 24543557 ---CUUUUUUCGUUUUUCGACAUACCUU-UCUUCUUAUUCG---CGCUGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAA ---...(((((((...(((((.......-.....((((((.---(((((.....)))))......((...))...............)))))).((((....))))...)))))...))))))) ( -24.60, z-score = -1.46, R) >droSim1.chr3L 9638829 117 - 22553184 CCUUUUUUCUCGUUUUUCGACAUACCU----UUCUUAUUCA---CUCUGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUAGUCUGCCGUUGAUUUGCGAAAA .........((((...(((((......----..........---............(((.....)))...(((......((((((((.......))))))))...))).)))))...))))... ( -19.40, z-score = -0.96, R) >droSec1.super_0 2463765 117 - 21120651 CCUUUUUUCUCGUUUUUCGACAUACCU----UUCUUAUUCA---CUCUGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAA .........((((...(((((......----......(((.---....)))...(((((((((((((...))..((((((...))))))......)))))))).)))..)))))...))))... ( -22.60, z-score = -1.97, R) >droYak2.chr3L 10236545 113 - 24197627 ----CUCUUUUUUUUUUCGACAUACC-U---UUCUUAUUCA---CUCCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAA ----.........((((((.((....-.---......(((.---....)))((((.(((.....)))...((..((((((...)))))).....((((....)))))).))))...)))))))) ( -17.40, z-score = -0.60, R) >droEre2.scaffold_4784 10240839 114 - 25762168 ----CUCUUCUUUUUUUCGACAUACCUU---UUCUUAUUCA---GUCCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAA ----.........((((((.((......---......(((.---....)))((((.(((.....)))...((..((((((...)))))).....((((....)))))).))))...)))))))) ( -17.40, z-score = -0.17, R) >droAna3.scaffold_13337 15163274 107 - 23293914 ------------UCUGUCAGCAUACCU----UUCCUAUUCA-UUCCCCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGCUGAUUUGCGAAAA ------------...((((((......----......(((.-......)))...(((((((((((((...))..((((((...))))))......)))))))).)))..))))))......... ( -20.20, z-score = -1.99, R) >droPer1.super_27 1085140 112 - 1201981 ---CUCUUUCUCUUUCUCUACCU--------UUCUUAUUUA-CUCGGCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAA ---....................--------..........-....(((((((((.(((.....)))...((..((((((...)))))).....((((....)))))).)))).)))))..... ( -18.80, z-score = -0.88, R) >droWil1.scaffold_181024 1369637 120 + 1751709 ---CUGUGUGUGCUCGUCGGUGUGUCUCCCCUUUUCAUUCA-CUCUCGGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAA ---((((.(((..(((..((((.................))-))..))).)))))))......(((((((.(((((((((...)))))).....((((....))))....))).)))))))... ( -23.43, z-score = -0.06, R) >droMoj3.scaffold_6680 4013486 123 + 24764193 -GCUUCCUGCUCCCACGAAGUCUUUCUGUGCUACUUGCUCGACGACAACGACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGUCGCUGAUUUGCGAAAA -((.....)).....((..(((..((.(.((.....)).))).)))..))....(((((((((((((...))..((((((...))))))......)))))))).)))((((......))))... ( -25.10, z-score = -0.49, R) >droVir3.scaffold_13049 16026886 123 - 25233164 -CUUUCUCUCUCGCUCGCAGUAUUUCUCUGCUGCUUGCGCUACUUGCUUAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGUCGCUGAUUUGCGAAAA -...........((..((((((......))))))..))((.....)).......(((((((((((((...))..((((((...))))))......)))))))).)))((((......))))... ( -30.10, z-score = -1.97, R) >droGri2.scaffold_15110 11929469 111 + 24565398 -UCUUUCUCAUCGCUCGAAUUCCUUCCCUGCUGUUUGCGCU------------ACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUCUUGUCUGUCGUUGAUUUGCGAAAA -..((((...((....))...........(((((.......------------))))))))).(((((((.(((((((((...)))))).....((((....))))....))).)))))))... ( -21.00, z-score = -0.71, R) >consensus ___UUUUUUCUCUUUUUCGACAUACCU____UUCUUAUUCA___CUCCGAACAACAGCGAAAAUCGCAAAGCAAUCAAAUAAAAUUUGAAUAAAAGAUUUUUGUCUGCCGUUGAUUUGCGAAAA ......................................................(((((((((((((...))..((((((...))))))......)))))))).))).(((......))).... (-14.28 = -14.26 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:31 2011