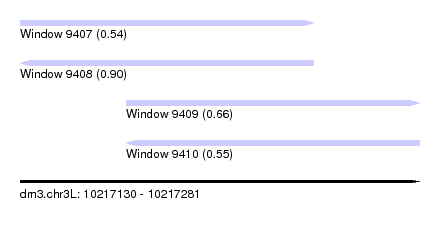
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,217,130 – 10,217,281 |
| Length | 151 |
| Max. P | 0.896756 |

| Location | 10,217,130 – 10,217,241 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.82 |
| Shannon entropy | 0.55172 |
| G+C content | 0.64021 |
| Mean single sequence MFE | -47.44 |
| Consensus MFE | -21.96 |
| Energy contribution | -24.20 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10217130 111 + 24543557 CGGUGGUGCUCCUCCUCCGUAGUGUGGCUGUCCACCAGGACCACCUACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUAAGGUGGCUGACCACCUGGACCACCCAUGUA .((((((.(............((((((..(((((...(((((((((.....))))))...)))...)))))...)))))).((((((....))))))).))))))...... ( -48.00, z-score = -1.82, R) >droSim1.chr3L 9606247 111 + 22553184 UGGUGGUGCUCCUCCUCCGUAGGGCGGCUGUCCACCAGGACCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGUCCACCCAUGUA .((((((.(....((((((((..((((..(((((...((((...((((.....))))..))))...)))))...)))).))))))))..)))))))(((.....))).... ( -50.70, z-score = -0.97, R) >droSec1.super_0 2432437 111 + 21120651 UGGUGGUGCUCCUCCUCCGUAGGGCGGCUGUCCACCAGGACCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGACCACCCAUGUA .((((((..((..((((((((..((((..(((((...((((...((((.....))))..))))...)))))...)))).))))))))..))((....))))))))...... ( -51.40, z-score = -1.27, R) >droEre2.scaffold_4784 10205976 99 + 25762168 AGGUGGUGCUCCUCCUCCGUAGGGUGGCUGUCCACCGGGACCACCUCCAUAAGGUGG------------AGCACCCGCAUAAGGUGGCUGGCCACCUGGACCACCCACGUA .(((((((((((.(((.....((((((...(((....))))))))).....))).))------------))))))......((((((....))))))..)))......... ( -45.90, z-score = -1.08, R) >droGri2.scaffold_15110 11899280 102 - 24565398 CGGCUGAGGUCCUGGUCCAUAUGCUUGCGGCGGACCAGCUGGUCCUGGCGGAUACUCAAAUGAAUGUGGAGCACCACUACCAGC--GCCAGC-GCCUGCGCCAAA------ .(((..((((..((((((...((....))..))))))((((((.((((.......((....))..((((....))))..)))).--))))))-))))..)))...------ ( -41.20, z-score = -1.02, R) >consensus CGGUGGUGCUCCUCCUCCGUAGGGUGGCUGUCCACCAGGACCACCUACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUAAGGGGGCUGACCACCUGGACCACCCAUGUA .((((((...((.(((.....((((((..((((....))))..........((((((....))))))...)))))).....))).))...))))))............... (-21.96 = -24.20 + 2.24)



| Location | 10,217,130 – 10,217,241 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.82 |
| Shannon entropy | 0.55172 |
| G+C content | 0.64021 |
| Mean single sequence MFE | -49.80 |
| Consensus MFE | -28.84 |
| Energy contribution | -31.56 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10217130 111 - 24543557 UACAUGGGUGGUCCAGGUGGUCAGCCACCUUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUAGGUGGUCCUGGUGGACAGCCACACUACGGAGGAGGAGCACCACCG ......((((((..((((((....))))))...((.(((.((((....)))).))).((((.((((((((((((....))...))))).)))))...)))).)))))))). ( -51.20, z-score = -1.69, R) >droSim1.chr3L 9606247 111 - 22553184 UACAUGGGUGGACCAGGUGGUCAGCCCCCGUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUGGGUGGUCCUGGUGGACAGCCGCCCUACGGAGGAGGAGCACCACCA ((((((((((((((((((.....)))((((....)))))))))))(((.....)))..))))))))((((.(((((((((....)))))(....)...)))).)))).... ( -53.30, z-score = -0.83, R) >droSec1.super_0 2432437 111 - 21120651 UACAUGGGUGGUCCAGGUGGUCAGCCCCCGUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUGGGUGGUCCUGGUGGACAGCCGCCCUACGGAGGAGGAGCACCACCA ......((((.(((.(((.(((.(((((((....)))).)))(((((......(((((((.....))))))))))))..))).))).(((....).)).))).)))).... ( -52.50, z-score = -0.52, R) >droEre2.scaffold_4784 10205976 99 - 25762168 UACGUGGGUGGUCCAGGUGGCCAGCCACCUUAUGCGGGUGCU------------CCACCUUAUGGAGGUGGUCCCGGUGGACAGCCACCCUACGGAGGAGGAGCACCACCU ...((((.(((((.....))))).)))).....(..((((((------------((.((((.(((.((((((((....))...)))))))))..)))).))))))))..). ( -48.90, z-score = -1.59, R) >droGri2.scaffold_15110 11899280 102 + 24565398 ------UUUGGCGCAGGC-GCUGGC--GCUGGUAGUGGUGCUCCACAUUCAUUUGAGUAUCCGCCAGGACCAGCUGGUCCGCCGCAAGCAUAUGGACCAGGACCUCAGCCG ------...(((..((((-((((((--.(((((.((((....))))((((....))))....)))))).)))))(((((((.(....)....))))))).).)))..))). ( -43.10, z-score = -1.58, R) >consensus UACAUGGGUGGUCCAGGUGGUCAGCCACCUUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUAGGUGGUCCUGGUGGACAGCCACCCUACGGAGGAGGAGCACCACCA ....(((.....)))((((((...((.((((....((((((((((((......(((((((.....))))))))))))......)))))))....)))).))...)))))). (-28.84 = -31.56 + 2.72)



| Location | 10,217,170 – 10,217,281 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.73 |
| Shannon entropy | 0.55283 |
| G+C content | 0.62015 |
| Mean single sequence MFE | -42.92 |
| Consensus MFE | -22.36 |
| Energy contribution | -26.20 |
| Covariance contribution | 3.84 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10217170 111 + 24543557 CCACCUACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUAAGGUGGCUGACCACCUGGACCACCCAUGUAAGGUGGUUGUCCGGCUCCGUAGGGCGGAGCUUCUCCGUAU ...(((((...((((((....))))))(((((.........((((((....)))))).(((((((.......)))))))....)).))))))))((((((...)))))).. ( -49.30, z-score = -2.15, R) >droSim1.chr3L 9606287 111 + 22553184 CCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGUCCACCCAUGUAAGGUGGUUGUCCGGCCCCGUAGGGCGGAGCUUCUCCGUAC (((((.......)))))...(((((((((.....))...(((((((.((((((((((((....))......)))))))))...).))))))))))))))............ ( -46.30, z-score = -0.10, R) >droSec1.super_0 2432477 111 + 21120651 CCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGACCACCCAUGUAAGGUGGUUGUCCGGCUCCGUAGGGCGGAGCUUCUCCGUAC (((((.......)))))..((((....))))........(((((((((.((((((((..((.......)).))))))))))))(((((((.....)))))))...))))). ( -47.40, z-score = -0.69, R) >droEre2.scaffold_4784 10206016 99 + 25762168 CCACCUCCAUAAGGU------------GGAGCACCCGCAUAAGGUGGCUGGCCACCUGGACCACCCACGUAAGGUGGUUGUCCGGCUCCGUAGGGCUGAGCUUCUCCGUAC ((((((.....))))------------))(((.((..(...((((((....))))))((((.(((..(....)..))).))))......)..))))).............. ( -39.40, z-score = -0.71, R) >droGri2.scaffold_15110 11899320 99 - 24565398 --GGUCCUGGCGGAUACUCAAAUGAAUGUGGAGCACCACUACCAGCGCCAGC-GCCUGCGCCA------AAAGGUGCUGAUCCACCGCCAUAA---UAGACAUCUCCGUAA --.(((.((((((..............((((....))))...(((((((.((-(....)))..------...))))))).....))))))...---..))).......... ( -32.20, z-score = -1.79, R) >consensus CCACCCACAUAAGGUGGCUGUCCGCCUGGACCACCCGCAUACGGGGGCUGACCACCUGGACCACCCAUGUAAGGUGGUUGUCCGGCUCCGUAGGGCGGAGCUUCUCCGUAC ((((((.....))))))...(((((((((.....)).......((((((((((((((..............)))))))....)))))))...)))))))............ (-22.36 = -26.20 + 3.84)


| Location | 10,217,170 – 10,217,281 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.73 |
| Shannon entropy | 0.55283 |
| G+C content | 0.62015 |
| Mean single sequence MFE | -46.20 |
| Consensus MFE | -21.12 |
| Energy contribution | -23.64 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10217170 111 - 24543557 AUACGGAGAAGCUCCGCCCUACGGAGCCGGACAACCACCUUACAUGGGUGGUCCAGGUGGUCAGCCACCUUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUAGGUGG ..........((((((.....)))))).......((((((.(((((((((((((((((((....)))))).....))...((((....)))).)))))).))))))))))) ( -49.80, z-score = -1.98, R) >droSim1.chr3L 9606287 111 - 22553184 GUACGGAGAAGCUCCGCCCUACGGGGCCGGACAACCACCUUACAUGGGUGGACCAGGUGGUCAGCCCCCGUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUGGGUGG ((.(((((...))))((((....)))).).))..((((((.(((((((((((((((((.....)))((((....)))))))))))(((.....)))..))))))))))))) ( -55.60, z-score = -1.84, R) >droSec1.super_0 2432477 111 - 21120651 GUACGGAGAAGCUCCGCCCUACGGAGCCGGACAACCACCUUACAUGGGUGGUCCAGGUGGUCAGCCCCCGUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUGGGUGG ((.((.....((((((.....)))))))).))..((((((.(((((((((((((...(((...(((((((....)))).))))))...))...)))))).))))))))))) ( -48.60, z-score = -0.51, R) >droEre2.scaffold_4784 10206016 99 - 25762168 GUACGGAGAAGCUCAGCCCUACGGAGCCGGACAACCACCUUACGUGGGUGGUCCAGGUGGCCAGCCACCUUAUGCGGGUGCUCC------------ACCUUAUGGAGGUGG (((((((...(..(.((.(....).)).)..)..((((((.....)))))))))((((((....)))))).......)))).((------------(((((...))))))) ( -39.60, z-score = -0.42, R) >droGri2.scaffold_15110 11899320 99 + 24565398 UUACGGAGAUGUCUA---UUAUGGCGGUGGAUCAGCACCUUU------UGGCGCAGGC-GCUGGCGCUGGUAGUGGUGCUCCACAUUCAUUUGAGUAUCCGCCAGGACC-- ..........((((.---...((((((((((...(((((..(------..((((....-....))))..)....))))))))))((((....))))...))))))))).-- ( -37.40, z-score = -1.47, R) >consensus GUACGGAGAAGCUCCGCCCUACGGAGCCGGACAACCACCUUACAUGGGUGGUCCAGGUGGUCAGCCCCCGUAUGCGGGUGGUCCAGGCGGACAGCCACCUUAUGUGGGUGG ((((((....((((((.....)))))).(((...((((((.....))))))))).(((.....))).))))))..(((...)))..........((((((.....)))))) (-21.12 = -23.64 + 2.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:27 2011