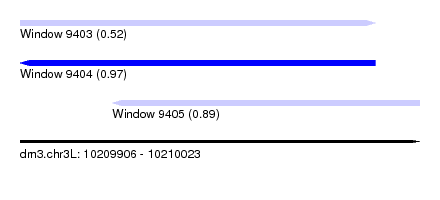
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,209,906 – 10,210,023 |
| Length | 117 |
| Max. P | 0.974517 |

| Location | 10,209,906 – 10,210,010 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.23 |
| Shannon entropy | 0.44795 |
| G+C content | 0.33236 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -9.65 |
| Energy contribution | -10.17 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518996 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
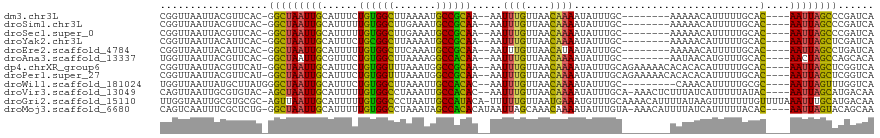
>dm3.chr3L 10209906 104 + 24543557 UGAUCGGGCUAAUU----GUGCAAAAAUGUUUUU--------GCAAAUAUUUUGUUAACAAAUU--UUGCGGCAUUUUAAGCCACAGAAAUGCAAUUAGCC-GUGAACGUAAUUAACCG ...(((((((((((----((((((((.((((...--------((((.....)))).))))..))--))))(((.......)))........))))))))))-.)))............. ( -29.10, z-score = -2.70, R) >droSim1.chr3L 9598884 104 + 22553184 UGAUCGGGCUAAUU----GUGCAAAAAUGUUUUU--------GCAAAUAUUUUGUUAACAAAUU--UUGCGGCAUUUCAAGCCACAAAAAUGCAAUUAGCC-GUGAACGUAAUUAACCG ...(((((((((((----((((((((.((((...--------((((.....)))).))))..))--))))(((.......)))........))))))))))-.)))............. ( -29.10, z-score = -2.73, R) >droSec1.super_0 2425280 104 + 21120651 UGAUCGGGCUAAUU----GUGCAAAAAUGUUUUU--------GCAAAUAUUUUGUUAACAAAUU--UUGCGGCAUUUCAAGCCACAAAAAUGCAAUUAGCC-GUGAACGUAAUUAACCG ...(((((((((((----((((((((.((((...--------((((.....)))).))))..))--))))(((.......)))........))))))))))-.)))............. ( -29.10, z-score = -2.73, R) >droYak2.chr3L 10195952 104 + 24197627 UGAUCGAGCUAAUU----GUGCAAAAAUGUUUUU--------GCAAAUAUUUUGUUAACAAAUU--UUGCGGCAUUUUAAGCCGCAGAAAUGCAAUUAGCC-GUGAAUGUAAUUAACCG ....((.(((((((----((((((((....))))--------))......((((....))))((--(((((((.......)))))))))..))))))))))-)................ ( -31.00, z-score = -3.54, R) >droEre2.scaffold_4784 10198353 104 + 25762168 UGAUCAGGCUAAUU----GUGCAAAAAUGUUUUU--------GCAAAUAUUAUGUUAACAAAUU--UUGCGGCAUUUGAAGCCACAAAAAUGCAAUUAGCC-GUGAAUGUAAUUAACCG ...(((((((((((----((((((((....))))--------))..................((--(((.(((.......))).)))))..))))))))))-.)))............. ( -29.50, z-score = -3.14, R) >droAna3.scaffold_13337 15118703 104 + 23293914 UGUGCUGGCUAGUU----GUGCAAACAUGUUAUU--------GCAAAUAUUUUGUUAACAAAUU--UUGUGGCCUUUUAAGCCACAGAAACGCAAUUAGCC-GUGAACGUAAUUAACCA .((..(((((((((----(((......(((((..--------((((.....)))))))))..((--(((((((.......))))))))).)))))))))))-)...))........... ( -33.10, z-score = -3.38, R) >dp4.chrXR_group6 8601398 112 + 13314419 UGACCGAGCUAAUU----GUGCAAAAAUGUGUGUGUUUUUCUGCAAAUAUUUUGUUAACAAAUU--UUGCGGCCAUUUAAACCACAGAAAUGCAAUUAGCC-AUGAACGUAAUUAACCG .....(.(((((((----(((((....)))(((.((((..(((((((...((((....)))).)--))))))......)))))))......))))))))))-................. ( -24.80, z-score = -1.48, R) >droPer1.super_27 1041458 112 + 1201981 UGACCGAGCUAAUU----GUGCAAAAAUGUGUGUGUUUUUCUGCAAAUAUUUUGUUAACAAAUU--UUGCGGCCAUUUAAACCACAGAAAUGCAAUUAGCC-AUGAACGUAAUUAACCG .....(.(((((((----(((((....)))(((.((((..(((((((...((((....)))).)--))))))......)))))))......))))))))))-................. ( -24.80, z-score = -1.48, R) >droWil1.scaffold_181024 1294641 104 - 1751709 UGACCAAACUAAUU----GCGCAAAAAUGUUUG---------GCAAAUAUUUUGUUAACAAAUU--GUGUGGCAAUUUAAGCCACAGAAAUGCAAUUAGCCCAUAAGCAUAAUUAACCA ........((((((----(((......((((.(---------((((.....)))))))))..((--.((((((.......)))))).)).))))))))).................... ( -24.10, z-score = -1.69, R) >droVir3.scaffold_13049 15986356 111 + 25233164 UUGUCAUGCUAAUU----GUAUAAAAAUGAUAAAGAGUUU-UGCAAAUAUUUUGUUAACAAAUU--GUGUGGCAAUUUAGGCCACAAAAAUGCAAUUAGCU-GUACACGCAAUUAACUG .(((((.(((((((----((((.....((((((((.(((.-....))).)))))))).......--.((((((.......))))))...))))))))))))-).)))............ ( -27.90, z-score = -2.02, R) >droGri2.scaffold_15110 11891751 117 - 24565398 UUGUCAUGCAAAUUUAAAACAAAAAAACUUAUAAAAUGUUUUGCAAACAUUUCAUUAACAAAAA-UGUAUGGCAAUUAGGGCCACAAAAAUGCAAUUAACU-GCGCACGCAAUUACCAA ((((((((((..(((......))).........(((((((.....)))))))............-))))))))))...((((.........))...(((.(-((....))).))))).. ( -16.60, z-score = 0.88, R) >droMoj3.scaffold_6680 3966945 113 - 24764193 UUGCUGUACUAAUU----GUGUAAAAAUGAUAAAAUGUUU-UACAAAUAUUUUGUUUGCUAAUUAUGUGUGGCUAUUUAGGCCACAAAAAUGCAAUUAGCC-CAGAGCGAAAUUGACUG ...(((..((((((----((((......(((((((((((.-....)))))))))))...........(((((((.....)))))))...))))))))))..-))).............. ( -29.30, z-score = -3.18, R) >consensus UGAUCGGGCUAAUU____GUGCAAAAAUGUUUUU________GCAAAUAUUUUGUUAACAAAUU__UUGCGGCAAUUUAAGCCACAAAAAUGCAAUUAGCC_GUGAACGUAAUUAACCG .......(((((((............................(((.....((((....)))).....)))(((.......)))..........)))))))................... ( -9.65 = -10.17 + 0.52)

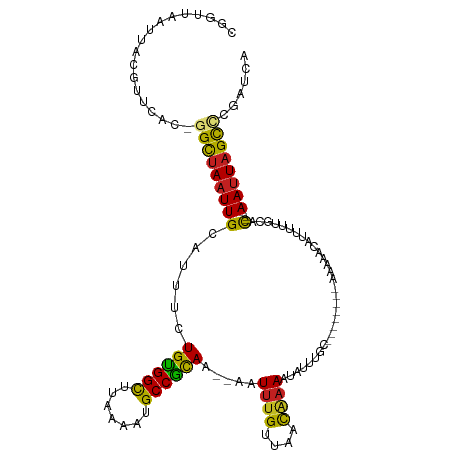
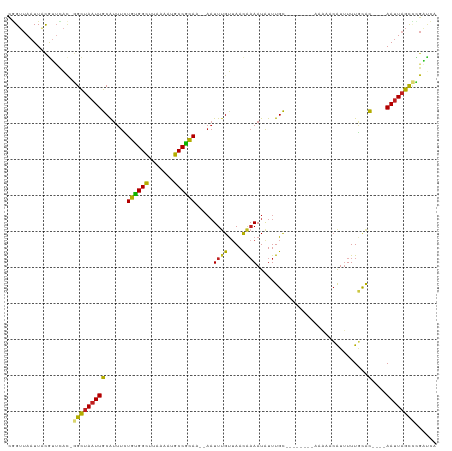
| Location | 10,209,906 – 10,210,010 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.23 |
| Shannon entropy | 0.44795 |
| G+C content | 0.33236 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -15.91 |
| Energy contribution | -15.47 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974517 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 10209906 104 - 24543557 CGGUUAAUUACGUUCAC-GGCUAAUUGCAUUUCUGUGGCUUAAAAUGCCGCAA--AAUUUGUUAACAAAAUAUUUGC--------AAAAACAUUUUUGCAC----AAUUAGCCCGAUCA .((((......(....)-(((((((((......((((((.......)))))).--..((((....)))).....(((--------((((....))))))))----)))))))).)))). ( -27.70, z-score = -2.99, R) >droSim1.chr3L 9598884 104 - 22553184 CGGUUAAUUACGUUCAC-GGCUAAUUGCAUUUUUGUGGCUUGAAAUGCCGCAA--AAUUUGUUAACAAAAUAUUUGC--------AAAAACAUUUUUGCAC----AAUUAGCCCGAUCA .((((......(....)-(((((((((...(((((((((.......)))))))--))((((....)))).....(((--------((((....))))))))----)))))))).)))). ( -30.30, z-score = -3.34, R) >droSec1.super_0 2425280 104 - 21120651 CGGUUAAUUACGUUCAC-GGCUAAUUGCAUUUUUGUGGCUUGAAAUGCCGCAA--AAUUUGUUAACAAAAUAUUUGC--------AAAAACAUUUUUGCAC----AAUUAGCCCGAUCA .((((......(....)-(((((((((...(((((((((.......)))))))--))((((....)))).....(((--------((((....))))))))----)))))))).)))). ( -30.30, z-score = -3.34, R) >droYak2.chr3L 10195952 104 - 24197627 CGGUUAAUUACAUUCAC-GGCUAAUUGCAUUUCUGCGGCUUAAAAUGCCGCAA--AAUUUGUUAACAAAAUAUUUGC--------AAAAACAUUUUUGCAC----AAUUAGCUCGAUCA ................(-(((((((((......((((((.......)))))).--..((((....)))).....(((--------((((....))))))))----))))))).)).... ( -28.30, z-score = -3.90, R) >droEre2.scaffold_4784 10198353 104 - 25762168 CGGUUAAUUACAUUCAC-GGCUAAUUGCAUUUUUGUGGCUUCAAAUGCCGCAA--AAUUUGUUAACAUAAUAUUUGC--------AAAAACAUUUUUGCAC----AAUUAGCCUGAUCA .............(((.-(((((((((...(((((((((.......)))))))--)).................(((--------((((....))))))))----)))))))))))... ( -31.50, z-score = -4.58, R) >droAna3.scaffold_13337 15118703 104 - 23293914 UGGUUAAUUACGUUCAC-GGCUAAUUGCGUUUCUGUGGCUUAAAAGGCCACAA--AAUUUGUUAACAAAAUAUUUGC--------AAUAACAUGUUUGCAC----AACUAGCCAGCACA ((((((.....(((.((-(((.....)).....(((((((.....))))))).--....))).)))........(((--------((........))))).----...))))))..... ( -25.10, z-score = -1.27, R) >dp4.chrXR_group6 8601398 112 - 13314419 CGGUUAAUUACGUUCAU-GGCUAAUUGCAUUUCUGUGGUUUAAAUGGCCGCAA--AAUUUGUUAACAAAAUAUUUGCAGAAAAACACACACAUUUUUGCAC----AAUUAGCUCGGUCA .................-(((((((((......(((((((.....))))))).--..((((....)))).....((((((((..........)))))))))----))))))))...... ( -23.60, z-score = -0.98, R) >droPer1.super_27 1041458 112 - 1201981 CGGUUAAUUACGUUCAU-GGCUAAUUGCAUUUCUGUGGUUUAAAUGGCCGCAA--AAUUUGUUAACAAAAUAUUUGCAGAAAAACACACACAUUUUUGCAC----AAUUAGCUCGGUCA .................-(((((((((......(((((((.....))))))).--..((((....)))).....((((((((..........)))))))))----))))))))...... ( -23.60, z-score = -0.98, R) >droWil1.scaffold_181024 1294641 104 + 1751709 UGGUUAAUUAUGCUUAUGGGCUAAUUGCAUUUCUGUGGCUUAAAUUGCCACAC--AAUUUGUUAACAAAAUAUUUGC---------CAAACAUUUUUGCGC----AAUUAGUUUGGUCA ................(..(((((((((.....((((((.......))))))(--((..((((..(((.....))).---------..))))...))).))----)))))))..).... ( -24.70, z-score = -1.03, R) >droVir3.scaffold_13049 15986356 111 - 25233164 CAGUUAAUUGCGUGUAC-AGCUAAUUGCAUUUUUGUGGCCUAAAUUGCCACAC--AAUUUGUUAACAAAAUAUUUGCA-AAACUCUUUAUCAUUUUUAUAC----AAUUAGCAUGACAA ............(((.(-(((((((((......((((((.......)))))).--..(((((.............)))-))...................)----))))))).))))). ( -21.12, z-score = -0.63, R) >droGri2.scaffold_15110 11891751 117 + 24565398 UUGGUAAUUGCGUGCGC-AGUUAAUUGCAUUUUUGUGGCCCUAAUUGCCAUACA-UUUUUGUUAAUGAAAUGUUUGCAAAACAUUUUAUAAGUUUUUUUGUUUUAAAUUUGCAUGACAA ....(((((((....))-)))))..((((..((((((((.......)))))...-.........((((((((((.....))))))))))...............)))..))))...... ( -23.70, z-score = 0.08, R) >droMoj3.scaffold_6680 3966945 113 + 24764193 CAGUCAAUUUCGCUCUG-GGCUAAUUGCAUUUUUGUGGCCUAAAUAGCCACACAUAAUUAGCAAACAAAAUAUUUGUA-AAACAUUUUAUCAUUUUUACAC----AAUUAGUACAGCAA ...........(((...-.((((((((......((((((.......))))))......................((((-(((...........))))))))----)))))))..))).. ( -21.90, z-score = -2.13, R) >consensus CGGUUAAUUACGUUCAC_GGCUAAUUGCAUUUCUGUGGCUUAAAAUGCCGCAA__AAUUUGUUAACAAAAUAUUUGC________AAAAACAUUUUUGCAC____AAUUAGCCCGAUCA ..................((((((((.......((((((.......)))))).....((((....))))....................................))))))))...... (-15.91 = -15.47 + -0.45)
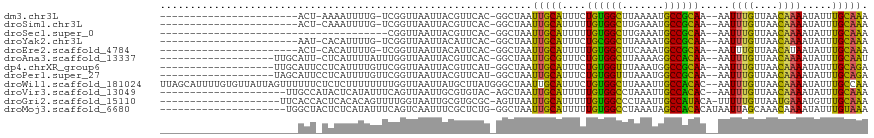
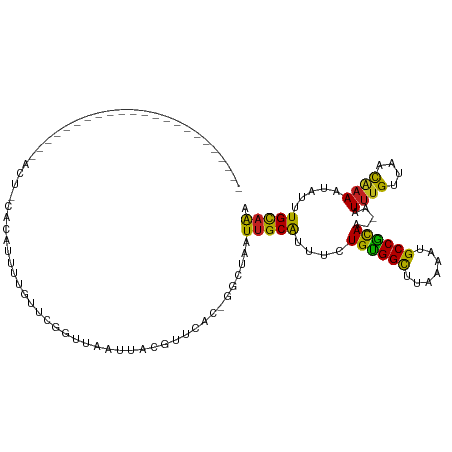
| Location | 10,209,933 – 10,210,023 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.02 |
| Shannon entropy | 0.50006 |
| G+C content | 0.32582 |
| Mean single sequence MFE | -18.98 |
| Consensus MFE | -13.64 |
| Energy contribution | -12.92 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.886601 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 10209933 90 - 24543557 -----------------------ACU-AAAAUUUUG-UCGGUUAAUUACGUUCAC-GGCUAAUUGCAUUUCUGUGGCUUAAAAUGCCGCAA--AAUUUGUUAACAAAAUAUUUGCAAA -----------------------...-........(-(((..............)-)))...(((((....((((((.......)))))).--..((((....)))).....))))). ( -17.94, z-score = -1.34, R) >droSim1.chr3L 9598911 90 - 22553184 -----------------------ACU-CAAAUUUUG-UCGGUUAAUUACGUUCAC-GGCUAAUUGCAUUUUUGUGGCUUGAAAUGCCGCAA--AAUUUGUUAACAAAAUAUUUGCAAA -----------------------...-........(-(((..............)-)))...(((((.(((((((((.......)))))))--))((((....)))).....))))). ( -20.54, z-score = -1.73, R) >droSec1.super_0 2425307 77 - 21120651 --------------------------------------CGGUUAAUUACGUUCAC-GGCUAAUUGCAUUUUUGUGGCUUGAAAUGCCGCAA--AAUUUGUUAACAAAAUAUUUGCAAA --------------------------------------..........((....)-).....(((((.(((((((((.......)))))))--))((((....)))).....))))). ( -17.90, z-score = -1.31, R) >droYak2.chr3L 10195979 90 - 24197627 -----------------------AAU-CACAUUUUG-UCGGUUAAUUACAUUCAC-GGCUAAUUGCAUUUCUGCGGCUUAAAAUGCCGCAA--AAUUUGUUAACAAAAUAUUUGCAAA -----------------------...-........(-(((..............)-)))...(((((....((((((.......)))))).--..((((....)))).....))))). ( -19.84, z-score = -2.03, R) >droEre2.scaffold_4784 10198380 90 - 25762168 -----------------------ACU-CACAUUUUG-UCGGUUAAUUACAUUCAC-GGCUAAUUGCAUUUUUGUGGCUUCAAAUGCCGCAA--AAUUUGUUAACAUAAUAUUUGCAAA -----------------------...-........(-(((..............)-)))...(((((.(((((((((.......)))))))--))..((....)).......))))). ( -18.44, z-score = -1.66, R) >droAna3.scaffold_13337 15118730 95 - 23293914 -------------------UUGCAUU-CUCAUUUUAUUUGGUUAAUUACGUUCAC-GGCUAAUUGCGUUUCUGUGGCUUAAAAGGCCACAA--AAUUUGUUAACAAAAUAUUUGCAAU -------------------(((((..-.......(((((.((((((..((....)-)((.....)).....(((((((.....))))))).--.....)))))).)))))..))))). ( -20.20, z-score = -1.85, R) >dp4.chrXR_group6 8601433 96 - 13314419 -------------------UUGCAUUCCUCAUUUUGUUCGGUUAAUUACGUUCAU-GGCUAAUUGCAUUUCUGUGGUUUAAAUGGCCGCAA--AAUUUGUUAACAAAAUAUUUGCAGA -------------------(((((......((((((((.((..((((........-.((.....)).....(((((((.....))))))).--))))..))))))))))...))))). ( -17.50, z-score = -0.09, R) >droPer1.super_27 1041493 96 - 1201981 -------------------UAGCAUUCCUCAUUUUGUUCGGUUAAUUACGUUCAU-GGCUAAUUGCAUUUCUGUGGUUUAAAUGGCCGCAA--AAUUUGUUAACAAAAUAUUUGCAGA -------------------((((...........((..((........))..)).-.)))).(((((....(((((((.....))))))).--..((((....)))).....))))). ( -17.42, z-score = -0.15, R) >droWil1.scaffold_181024 1294667 116 + 1751709 UUAGCAUUUUGUGUUAUUAGUUUUUUCUCUCUUUUUUUUGGUUAAUUAUGCUUAUGGGCUAAUUGCAUUUCUGUGGCUUAAAUUGCCACAC--AAUUUGUUAACAAAAUAUUUGCCAA ((((((..((((((.((((((((................(((.......)))...)))))))).))......(((((.......)))))))--))..))))))............... ( -21.41, z-score = -0.83, R) >droVir3.scaffold_13049 15986390 94 - 25233164 ---------------------UUGCCAUACUCAUAUUUCAGUUAAUUGCGUGUAC-AGCUAAUUGCAUUUUUGUGGCCUAAAUUGCCACAC--AAUUUGUUAACAAAAUAUUUGCAAA ---------------------((((.......((((((..((((((....(((..-.((.....))......(((((.......)))))))--)....)))))).))))))..)))). ( -17.80, z-score = -0.40, R) >droGri2.scaffold_15110 11891790 96 + 24565398 --------------------UUCACCACUCACACAGUUUUGGUAAUUGCGUGCGC-AGUUAAUUGCAUUUUUGUGGCCCUAAUUGCCAUACA-UUUUUGUUAAUGAAAUGUUUGCAAA --------------------....((((.(((.(((((.....))))).))).((-((....))))......))))......((((...(((-((((.......)))))))..)))). ( -21.40, z-score = -0.45, R) >droMoj3.scaffold_6680 3966979 96 + 24764193 ---------------------UGGCUACUCUCAUAUUUCAGUCAAUUUCGCUCUG-GGCUAAUUGCAUUUUUGUGGCCUAAAUAGCCACACAUAAUUAGCAAACAAAAUAUUUGUAAA ---------------------(((((.............)))))...........-.((((((((......((((((.......))))))..))))))))..((((.....))))... ( -17.32, z-score = -0.71, R) >consensus _______________________ACU_CACAUUUUGUUCGGUUAAUUACGUUCAC_GGCUAAUUGCAUUUCUGUGGCUUAAAAUGCCGCAA__AAUUUGUUAACAAAAUAUUUGCAAA ..............................................................(((((....((((((.......)))))).....((((....)))).....))))). (-13.64 = -12.92 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:23 2011