
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,194,309 – 10,194,469 |
| Length | 160 |
| Max. P | 0.592551 |

| Location | 10,194,309 – 10,194,469 |
|---|---|
| Length | 160 |
| Sequences | 6 |
| Columns | 177 |
| Reading direction | reverse |
| Mean pairwise identity | 64.84 |
| Shannon entropy | 0.66667 |
| G+C content | 0.31520 |
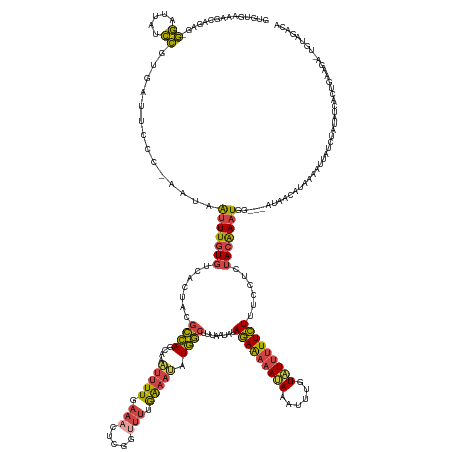
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -11.94 |
| Energy contribution | -11.38 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
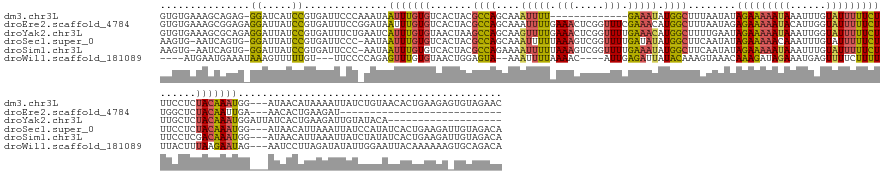
>dm3.chr3L 10194309 160 - 24543557 GUGUGAAAGCAGAG-GGAUCAUCCGUGAUUCCCAAAUAAUUUGUGUCACUACGCCAGCAAAUUUU-------------GAAAUAUGGCUUUAAUAUAGAAAAAUAAAUUUGUAUUUUUCUUUCCUCUACAAAUGG---AUAACAUAAAAUUAUCUGUAACACUGAAGAGUGUAGAAC ........((((((-((((((....))))))).......(((((((......((((.((.....)-------------).....))))........(((((((((......))))))))).(((.........))---)..)))))))....))))).(((((....)))))..... ( -32.90, z-score = -1.35, R) >droEre2.scaffold_4784 10182659 147 - 25762168 GUGUGAAAGCGGAGAGGAUUAUCCGUGAUUUCCGGAUAAUUUGUGUCACUACGCCAGCAAAUUUUGAAACUCGGUUUCGAAACAUGGCUUUAAUAGAGAAAAAUACAUUGGUAUUUUUCUUGGCUCUACAAUUGA---AACACUGAAGAU--------------------------- ((((...((.((...((((((((((.(....)))))))))))...)).))..((((((...(((((((((...)))))))))....)))......(((((((((((....))))))))))))))...........---.)))).......--------------------------- ( -41.10, z-score = -2.88, R) >droYak2.chr3L 10180005 158 - 24197627 GUGUGAAAGCGCAGAGGAUUAUCCGUGAUUUCUGAAUCAUUUGUGUAACUAAGCCAGCAAGUUUUGAAACUCGGUUUUGAAACAUGGCUUUUGAAUAGAAAAAUAAAUUGGUAUUUUUCUUUGCUCUACAAAUGGAUUAUCACUGAAGAUUGUAUACA------------------- ((((....))))..(.((((.((.(((((....((.(((((((((.....((((((....((((..((((...))))..)))).))))))..(...(((((((((......)))))))))...)..))))))))).))))))).)).)))).).....------------------- ( -40.10, z-score = -2.15, R) >droSec1.super_0 2409867 171 - 21120651 AAGUG-AAUCAGUG-GGAUUAUCCGUGAUUCCC-AAUAAUUUGUGUCACUACGCCAGCAAAUUUUUAAAGUCGGUUUUGAUAUAUGGCUUCAAUAUAGAAAAACAAAUUUGUAUUUUUCUUUCCUCUACAAAUGG---AUAACAUUAAAUUAUCCAUAUCACUGAAGAUUGUAGACA .((((-(..(((((-((((((....))).))))-).....)))..)))))......((((((((.........(((((..(((((.......)))))..)))))))))))))............(((((((((((---((((.......)))))))).((......))))))))).. ( -33.10, z-score = -0.65, R) >droSim1.chr3L 9583460 171 - 22553184 AAGUG-AAUCAGUG-GGAUUAUCCGUGAUUCCC-AAUAAUUUGUGUCACUACGCCAGAAAAUUUUUAAAGUCGGUUUUGAAAUAUGGCUUCAAUAUAGAAAAAUAAAUUUGUAUUUUUCUUUCCUCGACAAAUGG---AUAACAUUAAAUUAUCUAUAUCACUGAAGAUUGUAGACA .....-..((((((-(((......(((((...(-((....))).)))))...((((......((((((((....))))))))..))))........(((((((((......))))))))).))).......((((---((((.......))))))))..))))))............ ( -33.40, z-score = -0.71, R) >droWil1.scaffold_181089 5236098 161 - 12369635 ----AUGAAUGAAAUAAAGUUUUUGU---UUCCCCAGAGUUUGUGUAACUGGAGUA--AAAUUUUAAAAC----AUUGAGAUUAUACAAAGUAAACAAAGAUAGAAAUGAGUUUUCUUUUUUACUUUAAGAAUAG---AAUCCUUAGAUAUAUUGGAAUUACAAAAAAGUGCAGACA ----......(((((((.....))))---)))......(((((..(.......(((--.((((((((...----.)))))))).)))...((((.....((((....((((..((((.((((......)))).))---))..))))....))))....))))......)..))))). ( -27.70, z-score = -0.91, R) >consensus GUGUGAAAGCAGAG_GGAUUAUCCGUGAUUCCC_AAUAAUUUGUGUCACUACGCCAGCAAAUUUUGAAACUCGGUUUUGAAAUAUGGCUUUAAUAUAGAAAAAUAAAUUUGUAUUUUUCUUUCCUCUACAAAUGG___AUAACAUAAAAUUAUCUAUAUCACUGAAGA_UGUAGACA ...............((.....))..............(((((((.......((((....(((((.(((.....))).))))).))))........(((((((((......)))))))))......)))))))............................................ (-11.94 = -11.38 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:21 2011