| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,183,731 – 10,183,825 |
| Length | 94 |
| Max. P | 0.862494 |

| Location | 10,183,731 – 10,183,825 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.13 |
| Shannon entropy | 0.53523 |
| G+C content | 0.55467 |
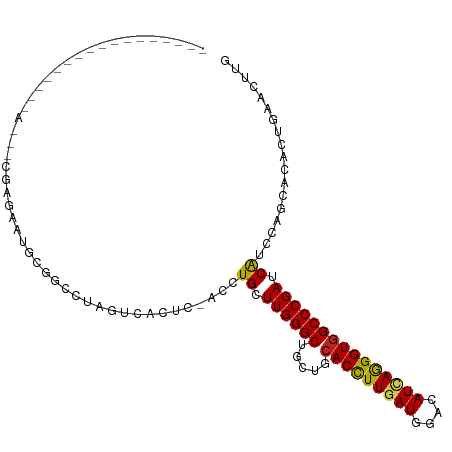
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.69 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
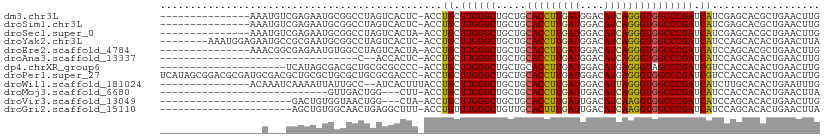
>dm3.chr3L 10183731 94 + 24543557 ---------------AAAUGUCGAGAAUGCGGCCUAGUCACUC-ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUCAGGGUGGCCCGAUCAUCGAGCACGCUGAACUUG ---------------......((((...(((((((((.((...-...)).)))))))))..(((((((((....)))))))))((.((.....)).))........)))) ( -32.50, z-score = -1.23, R) >droSim1.chr3L 9572919 94 + 22553184 ---------------AAAUGUCGAGAAUGCGGCCUAGUCACUC-ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUCAGGGUGGCCCGAUCAUCGAGCACGCUGAACUUG ---------------......((((...(((((((((.((...-...)).)))))))))..(((((((((....)))))))))((.((.....)).))........)))) ( -32.50, z-score = -1.23, R) >droSec1.super_0 2399256 94 + 21120651 ---------------AAAUGUCGAGAAUGCGGCCUAGUCACUA-ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUCAGGGUGGCCCGAUCAUCGAGCACGCUGAACUUG ---------------......((((...(((((((((.((...-...)).)))))))))..(((((((((....)))))))))((.((.....)).))........)))) ( -32.50, z-score = -1.62, R) >droYak2.chr3L 10168959 101 + 24197627 --------AAAUGGAGAAUGCCGCGAAUGCGGCCUAGUCACUC-ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUCAGGGUGGCCCGAUCAUCCAGCACACUGAACUUA --------...((((((..(((((....)))))....))....-...((.((((((.....(((((((((....))))))))))))))).)))))).............. ( -35.50, z-score = -1.46, R) >droEre2.scaffold_4784 10171811 94 + 25762168 ---------------AAACGGCGAGAAUGUGGCCUAGUCACUA-ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUCAGGGUGGCCCGAUCAUCCAGCACGCUGAACUUG ---------------...(((((.....(((((...)))))..-...((.((((((.....(((((((((....))))))))))))))).)).......)))))...... ( -33.10, z-score = -1.47, R) >droAna3.scaffold_13337 15093421 74 + 23293914 ---------------------------------C--ACCACUC-ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUCAGGGUGGCCCGAUCAUCCAGCACACUGAACUUG ---------------------------------.--.......-...((.((((((.....(((((((((....))))))))))))))).))..(((....)))...... ( -22.50, z-score = -1.07, R) >dp4.chrXR_group6 8576882 88 + 13314419 ---------------------UCAUAGCGACGCUGCGCGCCCC-ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUGAGGGUAGCCCGAUCGUCCACCACACUGAACUUG ---------------------...(((.(..((.(((.((((.-........))))))).))....((.(((((....(((...)))....))))).))).)))...... ( -24.50, z-score = 0.26, R) >droPer1.super_27 1016652 109 + 1201981 UCAUAGCGGACGCGAUGCGACGCUGCGCUGCGCUGCGCGACCC-ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUGAGGGUGGCCCGAUCGUCCACCACACUGAACUUG .....((((.((((.((((....)))).))))))))(((.(((-(......)))).))).......((.(((((....(((...)))....))))).))........... ( -34.60, z-score = 0.69, R) >droWil1.scaffold_181024 1259304 93 - 1751709 ---------------ACAAAUCAAAAUUAUUGCC--AUCACUUUACCUGCUUGGGCUGCUGCACCUUGAUGGACAUUAGGGUGGCCCGAUCAUCUUGCACACUGAAUUUG ---------------...................--...........((.((((((.....(((((((((....))))))))))))))).)).................. ( -19.20, z-score = 0.12, R) >droMoj3.scaffold_6680 3942029 78 - 24764193 ----------------------------GUUGACUGG---CUU-ACCUGCUUGGGCUGCUGCACCUUGAUUGACAUCAGGGUGGCCCGAUCAUCCACCACACUGAACUUA ----------------------------(((.(.(((---(..-....))((((((.....(((((((((....)))))))))))))))..........)).).)))... ( -22.80, z-score = -1.44, R) >droVir3.scaffold_13049 15962974 84 + 25233164 ----------------------GACUGUGGUAACUGG---CUA-ACCUGCUUGGGCUGCUGCACCUUGAUUGACAUCAAGGUGGCCCGAUCAUCCAGCACACUGAACUUG ----------------------....(((....((((---...-...((.((((((.....(((((((((....))))))))))))))).)).))))..)))........ ( -26.10, z-score = -1.63, R) >droGri2.scaffold_15110 11868352 87 - 24565398 ----------------------AGCUGUGGCAACUGAGGCUUU-ACCUGUUUGGGCUGUUGCACUUUGAUUGACAUCAAGGUGGCCCGAUCAUCCAGCACACUGAACUUA ----------------------.((((.((......(((....-.)))(.((((((.....(((((((((....))))))))))))))).).))))))............ ( -25.10, z-score = -0.38, R) >consensus _________________A___CGAGAAUGCGGCCUAGUCACUC_ACCUGCUUGGGCUGCUGCACCUUGAUGGACAUCAGGGUGGCCCGAUCAUCCAGCACACUGAACUUG ...............................................((.((((((.....(((((((((....))))))))))))))).)).................. (-19.86 = -19.69 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:17 2011