
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,181,400 – 10,181,497 |
| Length | 97 |
| Max. P | 0.892087 |

| Location | 10,181,400 – 10,181,497 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 64.30 |
| Shannon entropy | 0.75503 |
| G+C content | 0.38253 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -8.62 |
| Energy contribution | -8.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.87 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.892087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10181400 97 - 24543557 -UUCUGAUUGUAUUAC-AAUGUGGGCGGCAGCACGCCCCUGCCAUUUGAAAGUGGAGG-----AGAACA-GAAAUAUACGUAGAACAUUUUCAUAAAUCAUAAAA -(((((..((((((..-..((((((((......)))))((.(((((....))))).))-----...)))-..))))))..))))).................... ( -21.50, z-score = -0.97, R) >droSim1.chr3L 9570631 97 - 22553184 -UUCUGAUUGUAUUAC-AAUGUGGGCGGCAGCACGCCCCUUCCAUUCGAAAGUGGAGG-----AGAACA-GAAAUAUACGUAGAACAUUUUCAUAAAUCAUAAAA -(((((..((((((..-..((((((((......)))))(((((((......)))))))-----...)))-..))))))..))))).................... ( -25.50, z-score = -2.57, R) >droSec1.super_0 2396965 97 - 21120651 -UUCUGAUUGUAUUAC-AAUGUGGGCGGCAGCACGCCCCUUCCAUUUGAAAGUGGAGG-----AGAACA-GAAAUAUACGUAGAACAUUUUCAUAAAUCAUAAAA -(((((..((((((..-..((((((((......)))))((((((((....))))))))-----...)))-..))))))..))))).................... ( -25.60, z-score = -2.62, R) >droYak2.chr3L 10166596 102 - 24197627 -UCCUGAUUGUAUUAC-AAUGUGGGCGGCAUCACGCCCCUUUCAUUUGAAAGUGGAGGCGAACAGCACA-GAAAUAUACGUUGAACAUUUCCAUAAAUCAUAAAA -...(((((.......-.....(((((......)))))(((((....)))))((((((.(..((((...-.........))))..).))))))..)))))..... ( -22.40, z-score = -0.67, R) >droEre2.scaffold_4784 10169458 102 - 25762168 -UCCUGAUUGUAUUAC-AAUGUGGGCGGCAUCACGCCCCUUUCAUUUGAAAGUGGAGGAGCACAGAACA-GAAAUAUACGUAGAACAUUUCCAUAAAUCAUAAAA -...(((((.......-((((((((((......)))))((((((((....))))))))...........-..............)))))......)))))..... ( -21.22, z-score = -0.71, R) >dp4.chrXR_group6 8574652 96 - 13314419 -UCCUGAUUGUAAUAC-AAAGUGGGCGGAAUGACGCCCAACUGUUUAGCCAGGGGAG------AAGACA-GGAACAGAAAUAGAACAUUUUCAUAAAUCAUAAAA -(((((..........-....((((((......)))))).(((......))).....------....))-)))...((((........))))............. ( -22.40, z-score = -1.97, R) >droPer1.super_27 1014281 97 - 1201981 -UCCUGAUUGUAAUAC-AAAGUGGGCGGAAUGACGCCCAACUGUUUAGCCAGGGGAG------AAGACAGGGAACAGAAAUAGAACAUUUUCAUAAAUCAUAAAA -(((............-....((((((......)))))).((((((..((...))..------.)))))))))...((((........))))............. ( -22.00, z-score = -1.55, R) >droAna3.scaffold_13337 15091463 92 - 23293914 -UCCUGAUUGUAUUAC-AAUGUGGGCGGAAUGACG-CCCUCCUGUUU--AGCUGGA-------AAAACA-GAAGUAUUCGUAGAACAUUUUCAUAAAUCAUAAAA -...(((.(((.((((-.....(((((......))-)))..((((((--.......-------.)))))-)........)))).)))...)))............ ( -21.20, z-score = -1.69, R) >droWil1.scaffold_181024 1257328 94 + 1751709 GCCCUAAUUGCAUUAC-AAAGUGAGGCAAAAGAAGCAAGCCCUGUCUAAGGAAAGA-------AAUGAACGAAAAGGAUACCAAAUGU---AAUAAAUCAUAAAA ......((((((((..-...((...((.......))..))(((.....))).....-------....................)))))---)))........... ( -10.30, z-score = 0.56, R) >droMoj3.scaffold_6680 3939369 78 + 24764193 --GCUGAUUGCAUUACUCGGG-GGAGAUUCCAAUGUCCAAUUUAUUUGAGGAGA-------------CU--AAAAGUAAA---------CUCAUAAAUCAUAAUA --((.....)).......(((-(((.((....)).)))..(((((((.((....-------------))--..)))))))---------)))............. ( -12.90, z-score = -0.10, R) >droVir3.scaffold_13049 15961234 80 - 25233164 --GCUGAUUGCAUUACCAGAGUGGGUGGUCCACUGCCCUAUUUAUUCGACCAAA-------------CU-AAAAAAUAUU---------UUCAUAAAUCAUAAUA --((.....)).......(((((((..(....)..))).))))...........-------------..-..........---------................ ( -10.90, z-score = 0.19, R) >droGri2.scaffold_15110 11866778 84 + 24565398 --GCUGAUUGCAUUACCCCAGUGGGUGGUCCAAUGCCCUAUUUACUUGGAGGAA---------AAGUAU-GAAAAGGGAU------UCCUUCAUAAAUCAUA--- --..(((((((((((((((....)).)))..)))))..........((((((((---------.....(-....)....)------)))))))..)))))..--- ( -19.20, z-score = 0.25, R) >consensus _UCCUGAUUGUAUUAC_AAUGUGGGCGGCAUCACGCCCCUUUCAUUUGAAAGUGGAG______AAAACA_GAAAUAUACGUAGAACAUUUUCAUAAAUCAUAAAA ....(((((.............(((((......)))))...(((((....)))))........................................)))))..... ( -8.62 = -8.46 + -0.16)
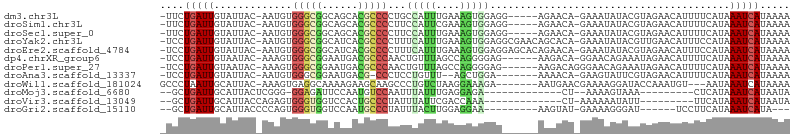

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:16 2011