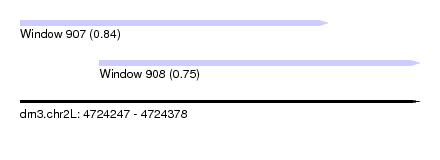
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,724,247 – 4,724,378 |
| Length | 131 |
| Max. P | 0.840359 |

| Location | 4,724,247 – 4,724,348 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 61.56 |
| Shannon entropy | 0.80060 |
| G+C content | 0.56749 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -16.88 |
| Energy contribution | -16.59 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.40 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
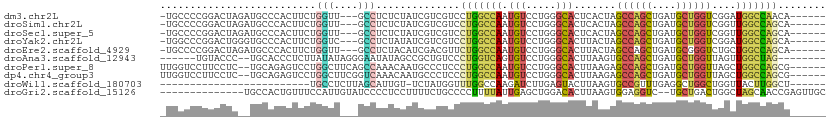
>dm3.chr2L 4724247 101 + 23011544 -UGCCCCGGACUAGAUGCCCACUUCUGGUU---GCCUCUCUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUCACUAGCCAGCUGAUGCUGGUCGGAUGGCCAACA------ -(((((.((((.(((.(((((....)))..---)).))).....(((.....)))....)))).))))).((((..((((((....))))))..).)))......------ ( -33.60, z-score = -0.37, R) >droSim1.chr2L 4592632 101 + 22036055 -UGCCCCGGACUAGAUGCCCACUUCUGGUU---GCCUCUCUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUCACUAGCCAGCUGAUGCUGGUCGGUUGGCCAGCA------ -(((((.((((.(((.(((((....)))..---)).))).....(((.....)))....)))).)))))....((.((((((((((....)))))))))).))..------ ( -36.40, z-score = -0.67, R) >droSec1.super_5 2800680 101 + 5866729 -UGCCCCGGACUAGAUGCCCACUUCUGGUU---GCCUCUCUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUCACUAGCCAGCUGAUGCUGGUCGGUUGGCCAGCA------ -(((((.((((.(((.(((((....)))..---)).))).....(((.....)))....)))).)))))....((.((((((((((....)))))))))).))..------ ( -36.40, z-score = -0.67, R) >droYak2.chr2L 4731839 101 + 22324452 -UGGCCCGGACUGGGUGCCCACUUCUGGUC---GCCUCUAUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUUACUAGCCAGCUGAUGCUGGUCGGAUGGCCAGCA------ -(((((.((((.(((((.(((....))).)---))))..........)))).))))).....((((.((....(..((((((....))))))..).)).))))..------ ( -41.80, z-score = -1.18, R) >droEre2.scaffold_4929 4799114 101 + 26641161 -UGCCCCGGACUAGAUGCCCACUUCUGGUU---GCCUCUACAUCGACGUUCUGGCCAAUGUCCUGGGCACUUACUAGCCAGCUGAUGCGGGUCUGCUGGCCAGCA------ -(((((.((((((((........))))(((---(((...((......))...)).)))))))).)))))....((.((((((.(((....))).)))))).))..------ ( -36.80, z-score = -0.84, R) >droAna3.scaffold_12943 1924715 95 - 5039921 ------UGUACCC--UGCACCCUCUUAUAUAGGGAAUAUAGCCGCUGUCCCUGGUCAGUGUCCUGGGCACUUAAGUGCCAGCUGAUGCUGGUUAGUUGGCUAG-------- ------....(((--((............)))))....(((((((((.((...((((((......(((((....))))).))))))...)).))).)))))).-------- ( -32.50, z-score = -0.82, R) >droPer1.super_8 1614900 103 - 3966273 UUGGUCCUUCCUC--UGCAGAGUCCUGGCUUCAGCCAAACAAUGCCCUCCCUGGCCAAUGUCCUGGGCACUUAAGAGCCAGCUGAUGCUGGUUAGCUGGCCAGCG------ ..((.....))..--.(((..((..((((....)))).))..))).....(((((((.(((.....)))......(((((((....)))))))...)))))))..------ ( -34.60, z-score = 0.12, R) >dp4.chr4_group3 570842 103 - 11692001 UUGGUCCUUCCUC--UGCAGAGUCCUGGCUUCGGUCAAACAAUGCCCUCCCUGGCCAAUGUCCUGGGCACUUAAGAGCCAGCUGAUGCUGGUUAGCUGGCCAGCG------ ..((.....))..--.(((..((..((((....)))).))..))).....(((((((.(((.....)))......(((((((....)))))))...)))))))..------ ( -35.00, z-score = -0.10, R) >droWil1.scaffold_180703 1462585 79 - 3946847 -------------------------UGCCUCUUAGCAUUGU-UCUAUGGUUUGGCCAAGAUCUUGAGUACUUAAGUGCCGUUUGAGGCUGGCUGGUUACUUGGCU------ -------------------------.(((((..(((.....-(((.(((.....))))))....(.((((....)))))))).))))).(((..(....)..)))------ ( -21.20, z-score = 0.07, R) >droGri2.scaffold_15126 3265618 95 - 8399593 --------------UGCCACUGUUUCCAUUGUAUCCCCUCCUUUUCUGCCCCUUUUAUUGAGCUGGACACUUAAGUGGAGGUC--UGCUGACUGGCUAGCAACCGAGUUGC --------------.(((......((((((........(((..(((.............)))..)))......))))))((((--....)))))))..(((((...))))) ( -20.86, z-score = 0.41, R) >consensus _UGCCCCGGACUA__UGCCCACUUCUGGUU___GCCUCUCUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUUAAGAGCCAGCUGAUGCUGGUCGGCUGGCCAGCA______ ..................................................(((((((.(((.....))).......((((((....))))))....)))))))........ (-16.88 = -16.59 + -0.29)

| Location | 4,724,273 – 4,724,378 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 56.72 |
| Shannon entropy | 0.92651 |
| G+C content | 0.55329 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.54 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.88 |
| Mean z-score | -0.51 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4724273 105 + 23011544 ---GUUGCCUCUCUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUCACUAGCCAGCUGAUGCUGGUCGGAUGGCCAACAGAUU-CCGGCAGACACAGUG-GGGCAGGUCCA---- ---.((((((((((..((..(((.((((((.(((.....)))....(..((((((....))))))..).))))))...))).-.))..)))......)-)))))).....---- ( -39.30, z-score = -0.21, R) >droSim1.chr2L 4592658 105 + 22036055 ---GUUGCCUCUCUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUCACUAGCCAGCUGAUGCUGGUCGGUUGGCCAGCAGAUU-CCGGCAGAAACAGUG-GGGCAGGUCCA---- ---.((((((((((..((..((((((((((.(((.....)))...((..((((((....))))))..)))))))))..))).-.))..)))......)-)))))).....---- ( -42.40, z-score = -1.09, R) >droSec1.super_5 2800706 105 + 5866729 ---GUUGCCUCUCUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUCACUAGCCAGCUGAUGCUGGUCGGUUGGCCAGCAGAUU-CCGGCAUACACAGUG-GGUCAGGUCCA---- ---.................(.(((((((..(((....(((........))).((((.((((((((....))))))))....-.))))....)))...-))))))).)..---- ( -37.40, z-score = -0.32, R) >droYak2.chr2L 4731865 105 + 22324452 ---GUCGCCUCUAUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUUACUAGCCAGCUGAUGCUGGUCGGAUGGCCAGCAGAAU-CCGCCAGACACAGUG-GGGCAGCUCCA---- ---((.(((.((((...((.(((.((((.....((((((.((....(..((((((....))))))..).)).))))..))..-..))))))))).)))-)))).))....---- ( -37.80, z-score = -0.08, R) >droEre2.scaffold_4929 4799140 105 + 26641161 ---GUUGCCUCUACAUCGACGUUCUGGCCAAUGUCCUGGGCACUUACUAGCCAGCUGAUGCGGGUCUGCUGGCCAGCAGACU-CCGGCAGACACAGUG-GGGCAGGUGCA---- ---.(((((((......((((((......))))))((((((........))).((((...((((((((((....))))))).-))).))).).))).)-)))))).....---- ( -40.00, z-score = 0.27, R) >droAna3.scaffold_12943 1924743 89 - 5039921 ---------UAUAGCCGCUGUCCCUGGUCAGUGUCCUGGGCACUUAAGUGCCAGCUGAUGCUGGUUAGUUGGCUAGGAAUAA------GGAUACAGUG-GAAUAG--------- ---------.....(((((((.(((........((((((.((((.....((((((....)))))).)).)).))))))...)------))..))))))-).....--------- ( -34.40, z-score = -2.30, R) >droPer1.super_8 1614925 108 - 3966273 GCUUCAGCCAAACAAUGCCCUCCCUGGCCAAUGUCCUGGGCACUUAAGAGCCAGCUGAUGCUGGUUAGCUGGCCAGCGAAAU-GGUUUCCAUACAAUG-GGGCACGUCUU---- ((....)).......(((((((.(((((((.(((.....)))......(((((((....)))))))...))))))).)).((-((...))))......-)))))......---- ( -37.00, z-score = -0.37, R) >dp4.chr4_group3 570867 108 - 11692001 GCUUCGGUCAAACAAUGCCCUCCCUGGCCAAUGUCCUGGGCACUUAAGAGCCAGCUGAUGCUGGUUAGCUGGCCAGCGAAAU-GGUUUCCAUACAAUG-GGGCACGUCUU---- ((....)).......(((((((.(((((((.(((.....)))......(((((((....)))))))...))))))).)).((-((...))))......-)))))......---- ( -38.00, z-score = -0.76, R) >droWil1.scaffold_180703 1462605 91 - 3946847 -------------------UGGUUUGGCCAAGAUCUUGAGUACUUAAGUGCCGUUUGAGGCUGGCUGGUUACUUGGCUUUUC-CUCUCAGUCCUAUAGUGGGUUGGUCUAA--- -------------------.((...(((((((..(((((....))))).(((......)))..........)))))))...)-)..(((..((......))..))).....--- ( -26.20, z-score = -0.40, R) >droMoj3.scaffold_6500 12100529 98 - 32352404 --------------CCCAUUGGCCAGAGCAUUUAAGUGCCAGUGUGCCAGCUGGCC--AGCUGUCCAGCAAGCCAGUUUGCUGCCUGAUAAGCCAAUUCACGGUGGUAAACAUA --------------....(((((((..((((....))))...)).))))).((((.--.(((....)))..))))((((((..(((((.........))).))..))))))... ( -33.90, z-score = -1.30, R) >droGri2.scaffold_15126 3265644 99 - 8399593 -------------CUUUUCUGCCCCUUUUAUUGAGCUGGACACUUAAGUGGAGGUC--UGCUGACUGGCUAGCAACCGAGUUGCUUGCUUAGCCCAUUCACAGUGGUCAAACUA -------------.......................((..((((...(((((((..--.((((...(((.((((((...)))))).))))))))).)))))))))..))..... ( -23.90, z-score = 0.94, R) >consensus ___GU_GCCUCUCUAUCGUCGUCCUGGCCAAUGUCCUGGGCACUUACGAGCCAGCUGAUGCUGGUCGGCUGGCCAGCAGAUU_CCGGCAGACACAGUG_GGGCAGGUCCA____ .......................(((((((.(((.....))).......((((((....))))))....)))))))...................................... (-14.53 = -14.54 + 0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:18 2011