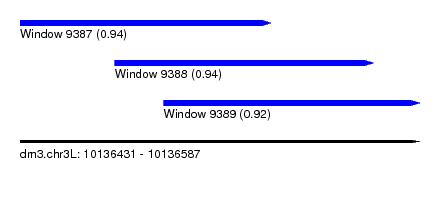
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,136,431 – 10,136,587 |
| Length | 156 |
| Max. P | 0.939104 |

| Location | 10,136,431 – 10,136,529 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 66.43 |
| Shannon entropy | 0.67024 |
| G+C content | 0.43566 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -10.17 |
| Energy contribution | -9.00 |
| Covariance contribution | -1.17 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10136431 98 + 24543557 ACUCCAUGGCUAUCCUUUCCGCCAGGGGAAUACGGGGCAAAUAGAUAGAGAGGCGGCCAAAGCAUUUGAUGUGUGUGUGCACAUACAUAUCAAGUGAA ......(((((..((((((.(((..(......)..))).........)))))).)))))...(((((((((((((((....))))))))))))))).. ( -35.80, z-score = -2.84, R) >droSim1.chr3L 9517663 98 + 22553184 ACUCUAUGGCCAUCCUUUUCGCCAGGGGAAUACGGGGCAAAUAGAUAGAGAGGCGGCCAAAGCAUUUGAUGUGUGUGUGCACAUACAUAUCAAGUGAA ...((.(((((..((((((.(((..(......)..))).........)))))).))))).))(((((((((((((((....))))))))))))))).. ( -36.30, z-score = -3.38, R) >droSec1.super_0 2352108 98 + 21120651 ACUCUAUGGCUAUCCUUUUCGCCAGGGGAAUACGGGGCAAAUAGAUAGAGAGUCGGCCAAAGCAUUUGAUGUGUGUGUGCACAUACAUAUAAAGUGAA (((((....(((((..(((.(((..(......)..))))))..)))))))))).........(((((.(((((((((....))))))))).))))).. ( -27.40, z-score = -1.20, R) >droEre2.scaffold_4784 10120492 98 + 25762168 ACUCCACGGCUAUCCUUUUCAGCAGGGGAGUACGGGGAAAACAGAUAGAGAGGCGGCCAAAGCAUUUGAUGUGUGUGUGCACAUACAUAUCAAGUGAA .((((...(((.(((((......))))))))..)))).........................(((((((((((((((....))))))))))))))).. ( -30.30, z-score = -1.90, R) >droWil1.scaffold_181024 1181011 79 - 1751709 ---------------UGCUGCCUGCUCUAAUACUCUGGCG-UGCAUAGA-AGGCGGCGAAAAUAUUUGAUAUGUAUGAGU--GUGCAUAUAAAGUGAA ---------------.(((((((..((((.(((......)-))..))))-))))))).....(((((.((((((((....--)))))))).))))).. ( -23.40, z-score = -2.09, R) >droVir3.scaffold_13049 15915603 89 + 25233164 GUUCCA-----AUGCUGAUCCUGCAGCGCCUUAUACGCUA-UGCAUAGA-AGACGGCGAAAAUAUUUGAUAUGUAUGAGU--GUGCAUAUAAAGUGAA ......-----.(((((.((.(((((((.......)))..-))))....-.)))))))....(((((.((((((((....--)))))))).))))).. ( -18.70, z-score = -0.03, R) >droMoj3.scaffold_6680 3877972 94 - 24764193 GUUCCAGAGGCAUUGGGAUGCCCCAGCUCCUUGUGCGCUA-UGCAUAGA-AGAGAGCUAAAAUAUUUUAUAUGUAUGAGU--GCGCAUAUAGAGUGAA ......(.(((((....))))).).((((..(((((((((-((((((.(-(((...........)))).))))))))...--)))))))..))))... ( -28.80, z-score = -2.20, R) >droGri2.scaffold_15110 11824716 89 - 24565398 AUUCCA-----UUCCUGAUUCUGUUGCGACUCUUAUGAUU-UACACAGA-AGGCGGCGAAAAUAUUUGAUAUGUAUGAGU--GUGCAUAUAAAGUGAA ......-----((((((.((((((.(..(.((....)).)-..))))))-)..))).)))..(((((.((((((((....--)))))))).))))).. ( -20.10, z-score = -1.25, R) >consensus ACUCCA_GGC_AUCCUGUUCCCCAGGCGAAUACGGGGCAA_UACAUAGA_AGGCGGCCAAAACAUUUGAUAUGUAUGAGC__AUACAUAUAAAGUGAA ..............................................................(((((.((((((((......)))))))).))))).. (-10.17 = -9.00 + -1.17)


| Location | 10,136,468 – 10,136,569 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 86.01 |
| Shannon entropy | 0.26051 |
| G+C content | 0.35053 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -18.33 |
| Energy contribution | -17.41 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10136468 101 + 24543557 CAAAUAGAUAGAGAGGCGGCCAAAGCAUUUGAUGU--GUGUGUGCACAUACAUAUCAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU .........................((((((((((--(((((....)))))))))))))))..........((((..(((((((....)))))))..)))).. ( -32.40, z-score = -3.52, R) >droSim1.chr3L 9517700 101 + 22553184 CAAAUAGAUAGAGAGGCGGCCAAAGCAUUUGAUGU--GUGUGUGCACAUACAUAUCAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU .........................((((((((((--(((((....)))))))))))))))..........((((..(((((((....)))))))..)))).. ( -32.40, z-score = -3.52, R) >droSec1.super_0 2352145 101 + 21120651 CAAAUAGAUAGAGAGUCGGCCAAAGCAUUUGAUGU--GUGUGUGCACAUACAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU ......(((.....)))........(((((.((((--(((((....))))))))).)))))..........((((..(((((((....)))))))..)))).. ( -26.80, z-score = -1.99, R) >droEre2.scaffold_4784 10120529 101 + 25762168 AAAACAGAUAGAGAGGCGGCCAAAGCAUUUGAUGU--GUGUGUGCACAUACAUAUCAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU .........................((((((((((--(((((....)))))))))))))))..........((((..(((((((....)))))))..)))).. ( -32.40, z-score = -3.45, R) >droAna3.scaffold_13337 15047603 103 + 23293914 AAAAUAGAUAGAGAGGCGGCCAAAGUAUUUGAUGUGCAUGUAUAAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU .........................(((((.(((((((..(....)..))))))).)))))..........((((..(((((((....)))))))..)))).. ( -23.80, z-score = -1.80, R) >droWil1.scaffold_181024 1181036 94 - 1751709 ----UGCAUAGA-AGGCGGCGAAAAUAUUUGAU----AUGUAUGAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU ----((((((..-..(((.((((....))))..----.))).....))))))......((....)).....((((..(((((((....)))))))..)))).. ( -22.30, z-score = -1.75, R) >droVir3.scaffold_13049 15915638 94 + 25233164 ----UGCAUAGA-AGACGGCGAAAAUAUUUGAU----AUGUAUGAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU ----((((((..-(((...........)))..)----)))))(((((((.(((.....)))...)))))))((((..(((((((....)))))))..)))).. ( -22.10, z-score = -1.83, R) >droMoj3.scaffold_6680 3878012 94 - 24764193 ----UGCAUAGA-AGAGAGCUAAAAUAUUUUAU----AUGUAUGAGUGCGCAUAUAGAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU ----........-............((((((((----((((........))))))))))))..........((((..(((((((....)))))))..)))).. ( -22.60, z-score = -2.15, R) >droGri2.scaffold_15110 11824751 94 - 24565398 ----UACACAGA-AGGCGGCGAAAAUAUUUGAU----AUGUAUGAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU ----((((((..-.(((..((.((((....(((----((((.(((((((.(((.....)))...))))))).)))))))..)))).)).)))...)))))).. ( -22.20, z-score = -1.73, R) >consensus _AAAUAGAUAGAGAGGCGGCCAAAGUAUUUGAUGU__AUGUAUGAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAU .....................................((((((....))))))...(((((....))))).((((..(((((((....)))))))..)))).. (-18.33 = -17.41 + -0.92)
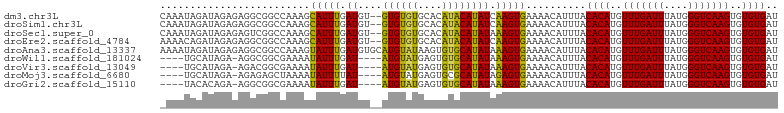
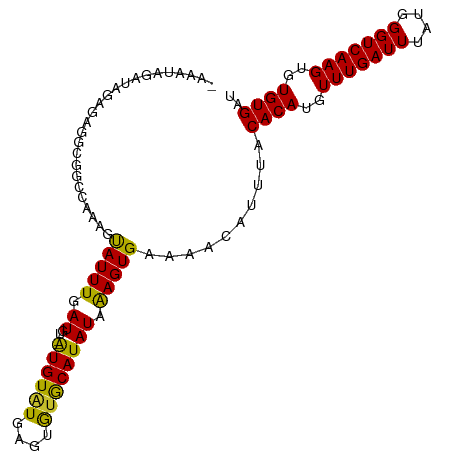
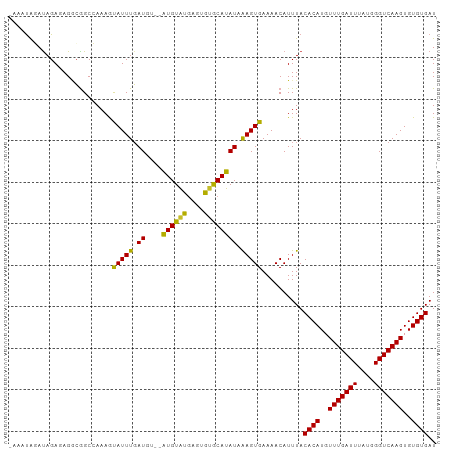
| Location | 10,136,487 – 10,136,587 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.90 |
| Shannon entropy | 0.20668 |
| G+C content | 0.33096 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -18.67 |
| Energy contribution | -17.84 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10136487 100 + 24543557 CCAAAGCAUUUGAUGU--GUGUGUGCACAUACAUAUCAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACAAAAGCA--GUC ......((((((((((--(((((....)))))))))))))))......((((((((..(((((((....)))))))..)))).))))............--... ( -32.90, z-score = -3.66, R) >droSim1.chr3L 9517719 100 + 22553184 CCAAAGCAUUUGAUGU--GUGUGUGCACAUACAUAUCAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACAAAAGCA--GUC ......((((((((((--(((((....)))))))))))))))......((((((((..(((((((....)))))))..)))).))))............--... ( -32.90, z-score = -3.66, R) >droSec1.super_0 2352164 100 + 21120651 CCAAAGCAUUUGAUGU--GUGUGUGCACAUACAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACAAAAGCU--GUC ....(((.((((((((--(((((....)))))))))...((....)).((((((((..(((((((....)))))))..)))).))))....)))).)))--... ( -29.50, z-score = -2.72, R) >droEre2.scaffold_4784 10120548 100 + 25762168 CCAAAGCAUUUGAUGU--GUGUGUGCACAUACAUAUCAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACAAAAGCA--GUC ......((((((((((--(((((....)))))))))))))))......((((((((..(((((((....)))))))..)))).))))............--... ( -32.90, z-score = -3.66, R) >droAna3.scaffold_13337 15047622 101 + 23293914 CCAAAGUAUUUGAUGUGCAUGUAUAAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUAC-AAAGCA--GUC .....((.(((((((((((..(....)..)))))))...((....)).((((((((..(((((((....)))))))..)))).))))....)-))))).--... ( -25.30, z-score = -1.90, R) >droWil1.scaffold_181024 1181050 98 - 1751709 CGAAAAUAUUUGAU----AUGUAUGAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACAAAAGCA--GCC ........((((((----((((((....))))))))...((....)).((((((((..(((((((....)))))))..)))).))))....))))....--... ( -22.20, z-score = -1.93, R) >droVir3.scaffold_13049 15915652 98 + 25233164 CGAAAAUAUUUGAU----AUGUAUGAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACAAAAGCA-AGC- ........((((((----((((((....))))))))...((....)).((((((((..(((((((....)))))))..)))).))))....))))....-...- ( -22.20, z-score = -2.10, R) >droMoj3.scaffold_6680 3878026 98 - 24764193 CUAAAAUAUUUUAU----AUGUAUGAGUGCGCAUAUAGAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACAAAAGCA-AGC- ......((((((((----((((........))))))))))))......((((((((..(((((((....)))))))..)))).))))............-...- ( -23.10, z-score = -2.23, R) >droGri2.scaffold_15110 11824765 99 - 24565398 CGAAAAUAUUUGAU----AUGUAUGAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACCAAAGCGGAGC- ......(((((.((----((((((....)))))))).)))))......((((((((..(((((((....)))))))..)))).))))....((.....))...- ( -22.70, z-score = -2.00, R) >consensus CCAAAGUAUUUGAUGU__AUGUAUGAGUGUGCAUAUAAAGUGAAAACAUUUACACAUGUUUGAUUUAUGGGUCAAGUGUGUGAUAAAACUACAAAAGCA__GUC ........(((.((....((((((....)))))).....)).)))...((((((((..(((((((....)))))))..)))).))))................. (-18.67 = -17.84 + -0.83)

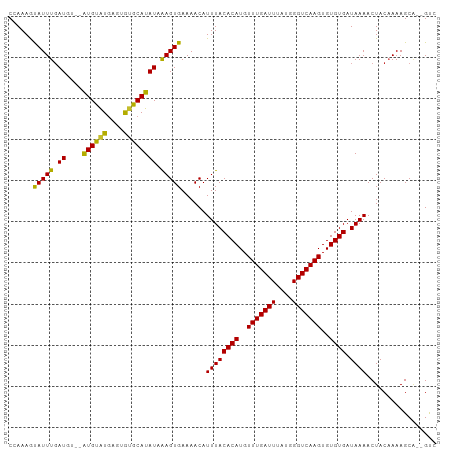
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:15:10 2011