| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,717,955 – 4,718,059 |
| Length | 104 |
| Max. P | 0.970031 |

| Location | 4,717,955 – 4,718,059 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.20 |
| Shannon entropy | 0.58765 |
| G+C content | 0.43072 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
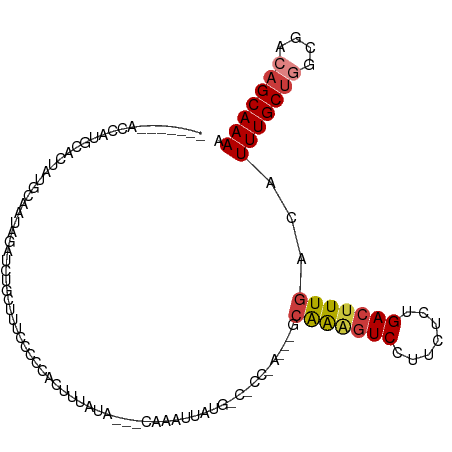
>dm3.chr2L 4717955 104 - 23011544 -------ACCAUGCACUAUGCAUUAGAUUUGCUUUCCCCCACUUUAUA---CAAAUUAUGGCACCCAAAAGCAGAGUCCUUCUCUGACUUUGACAUUUGCUGGCGACAGCAAAA -------.((((((.....))....((((((.................---))))))))))....((((..(((((.....)))))..))))...(((((((....))))))). ( -25.03, z-score = -2.85, R) >droSim1.chr2L 4586414 94 - 22036055 -------ACCAUGCACUAUGCAAUAGAUUUGCUUUCUCCCACUUUAGA---CAAAUUAUG----------GAGAAGUCCUUCUCUGACUUUGACAUUUGCUGGCGACAGCAAAA -------.((((((.....))....((((((((............)).---)))))))))----------).((((((.......))))))....(((((((....))))))). ( -22.50, z-score = -2.19, R) >droSec1.super_5 2794404 94 - 5866729 -------ACCAUGCACUAUGCAAUAGAUUUGCUUUCCCCCACUUUAGA---CAAAUUAUG----------GCAAAGUCCUUCUCUGACUUUGACAUUUGCUGGCGACAGCAAAA -------.((((((.....))....((((((((............)).---)))))))))----------)(((((((.......)))))))...(((((((....))))))). ( -24.30, z-score = -3.17, R) >droYak2.chr2L 4725453 102 - 22324452 -------ACCAUGCAC--UGCCAUAGAACUGAUUUCCCACACUUUAUA---CAAAUUAUGGCACCCAAAAGCAAAGUCCUUCGCUGAAAUUGACAUUUGCUGGCGACUGCAAAA -------....((((.--((((((((...((.................---))..))))))))((((...((((((((.(((...)))...))).)))))))).)..))))... ( -17.33, z-score = -0.33, R) >droEre2.scaffold_4929 4792584 92 - 26641161 -------ACCAUGCAC--UGGAACAGAACUGGUUUCCCGCACUUACUA---CAAAUUAUG----------GCAAAGUCCUUCGCUGACUUUGACAUUUGCUGGCGACAGCAAAA -------....(((..--.(((((.......)))))..))).......---.......((----------.(((((((.......))))))).))(((((((....))))))). ( -22.90, z-score = -1.67, R) >dp4.chr4_group3 564303 114 + 11692001 ACUGAAUCCAAUUGGCAACGAAAGCACCCAGUUUGUACCAAUUAUGUAGCCCCAAACGCAACUCCAAGUGCCAGGGUCCUUGUCUGACUUUGACAUUUGCUAACAACAGCAAAU ...........(((....)))(((.((((.((((((((.......)))....)))))(((........)))..)))).)))(((.......)))(((((((......))))))) ( -22.30, z-score = 0.20, R) >droPer1.super_8 1608300 114 + 3966273 ACUGAAUCCAAUUGGCAACGAAAGCACCCAGUUUGUACCAAUUAUGUAGCCCCAAACGCAACUCCAAGUGCCAGGGUCCUUGUCUGACUUUGACAUUUGCUAACAACAGCAAAU ...........(((....)))(((.((((.((((((((.......)))....)))))(((........)))..)))).)))(((.......)))(((((((......))))))) ( -22.30, z-score = 0.20, R) >consensus _______ACCAUGCACUAUGCAAUAGAUCUGCUUUCCCCCACUUUAUA___CAAAUUAUG_C_CC_A___GCAAAGUCCUUCUCUGACUUUGACAUUUGCUGGCGACAGCAAAA .......................................................................(((((((.......)))))))...(((((((....))))))). (-13.10 = -13.43 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:17 2011