
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,043,927 – 10,044,020 |
| Length | 93 |
| Max. P | 0.527996 |

| Location | 10,043,927 – 10,044,020 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Shannon entropy | 0.38945 |
| G+C content | 0.38441 |
| Mean single sequence MFE | -17.14 |
| Consensus MFE | -10.79 |
| Energy contribution | -10.26 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527996 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 10043927 93 + 24543557 ----UCAAACUCAAAGGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCGAACUUU-GGCGACCCGCUGGAAAUUACUUACUGCCAACUACAGAUACU--------------------- ----........((((((....)))))).......((((((((...((((......-..)).))...)))))))).....(((.......))).....--------------------- ( -16.00, z-score = -1.43, R) >droSim1.chr3L 9431887 93 + 22553184 ----UCAAACUCAAAGGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCGAACUUU-GGCGACCCGCUGGAAAUUACUUACUGCCAACUAAAGAUACU--------------------- ----........((((((....)))))).......((((((((...((((......-..)).))...)))))))).......................--------------------- ( -15.00, z-score = -0.93, R) >droSec1.super_0 2267163 93 + 21120651 ----UCAAACUCAAAGGCAUUUGCUCUUAAUUAAAAAUUUCCACUUGGCGAACUUU-GGCGACCCGCUGGAAAUUACUUACUGCCAACUACAGAUACU--------------------- ----...........((((((((........))))((((((((...((((......-..)).))...))))))))......)))).............--------------------- ( -14.70, z-score = -0.92, R) >droYak2.chr3L 10021942 93 + 24197627 ----UCAAACUCAAAGGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCGAACUUU-GGCGACCCGCUGCAAAUUACUUACUGCCAACUACAGAUACU--------------------- ----...........(((((((((.............((.((....)).)).....-((((...)))))))))........)))).............--------------------- ( -11.80, z-score = 0.16, R) >droEre2.scaffold_4784 10028855 93 + 25762168 ----UCAAACUCAAAGGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCGAACUUU-GGCGACCCGCUGGAAAUUACUUACUGCCAACUACAGAUACU--------------------- ----........((((((....)))))).......((((((((...((((......-..)).))...)))))))).....(((.......))).....--------------------- ( -16.00, z-score = -1.43, R) >droAna3.scaffold_13337 14961294 96 + 23293914 ACCAAAAAACUGAAAAGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCAAACUUUUGGAGACCCGCUGGAAAUUACUUACUGCCAACUACAGAUA----------------------- .........(((((((((....)))))).......((((((((..(((.........(....)))).))))))))...............)))...----------------------- ( -15.90, z-score = -1.62, R) >dp4.chrXR_group6 5712011 101 - 13314419 ----UCAAACUCAAAAGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCGAACUUU-GGGGACCAGCUGGAAAUUGCUUAUGCCCAAGUACAGAUACCCGACGCUC------------- ----........((((((....))))))..................((((....((-(((.(..(((........)))..).)))))(((....)))....)))).------------- ( -19.10, z-score = -0.13, R) >droPer1.super_27 854134 101 + 1201981 ----UCAAACUCAAAAGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCGAACUUU-GGGGACCAGCUGGAAAUUGCUUAUGCCCAAGUACAGAUACCGGACGCUC------------- ----........((((((....))))))..................((((..(.((-(((.(..(((........)))..).)))))(((....))).)..)))).------------- ( -19.50, z-score = -0.00, R) >droWil1.scaffold_180698 7705200 116 + 11422946 ---GUUAAACUCAAAAGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCGCACUUUUGGCGACCCACUGGAAAUUACUUACUGUCAUUUACAGCAACUUUGGCCCCCCGAUUCCCGAUC ---((((((...((((((....)))))).......((((((((...(((((.(....)))).))...)))))))).....((((.....))))....))))))................ ( -22.30, z-score = -1.59, R) >droVir3.scaffold_13049 9167206 83 + 25233164 ----UUAAACUUAAAAGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGCCGCACUUU-GCCGCCGCACUGGGAAUUACUUAC-AAAAACU------------------------------ ----......((((((((....)))))))).....((((((((..(((.((.....-...)).))).))))))))......-.......------------------------------ ( -19.10, z-score = -3.57, R) >droMoj3.scaffold_6680 18588048 79 - 24764193 ----UUAAACUUAAAAGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGCCGCACUUU-GCCGCCGCACUGGGAAUUAC-----AAAAACU------------------------------ ----......((((((((....)))))))).....((((((((..(((.((.....-...)).))).))))))))..-----.......------------------------------ ( -19.10, z-score = -3.33, R) >consensus ____UCAAACUCAAAAGCAUUUGCUUUUAAUUAAAAAUUUCCACUUGGCGAACUUU_GGCGACCCGCUGGAAAUUACUUACUGCCAACUACAGAUACU_____________________ ............((((((....)))))).......((((((((..(((...............))).))))))))............................................ (-10.79 = -10.26 + -0.53)

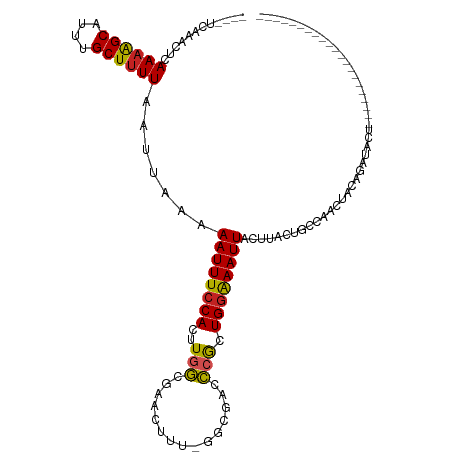

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:55 2011