| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,035,591 – 10,035,688 |
| Length | 97 |
| Max. P | 0.971502 |

| Location | 10,035,591 – 10,035,688 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.72 |
| Shannon entropy | 0.26415 |
| G+C content | 0.45729 |
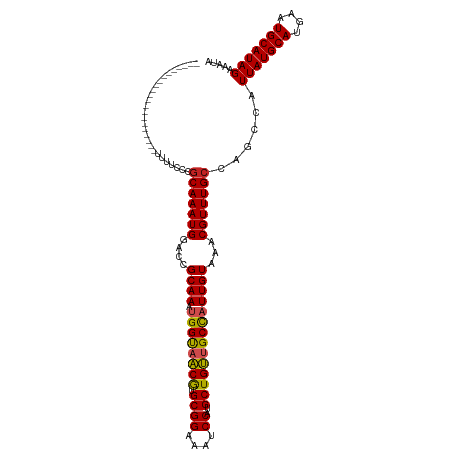
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -29.00 |
| Energy contribution | -28.73 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
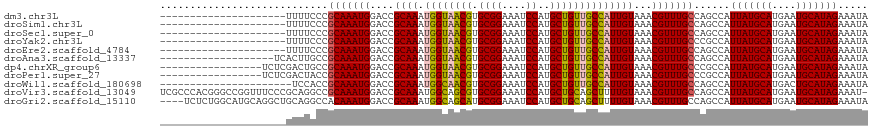
>dm3.chr3L 10035591 97 + 24543557 ---------------------UUUUCCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUA ---------------------.((((..(((((((....((((.((((((((.((((....))..))))))))))))))...))))))).......((((((....)))))))))).. ( -29.90, z-score = -2.21, R) >droSim1.chr3L 9423522 97 + 22553184 ---------------------UUUUCCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUA ---------------------.((((..(((((((....((((.((((((((.((((....))..))))))))))))))...))))))).......((((((....)))))))))).. ( -29.90, z-score = -2.21, R) >droSec1.super_0 2258900 97 + 21120651 ---------------------UUUUCCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUA ---------------------.((((..(((((((....((((.((((((((.((((....))..))))))))))))))...))))))).......((((((....)))))))))).. ( -29.90, z-score = -2.21, R) >droYak2.chr3L 10013446 97 + 24197627 ---------------------UUUUCCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCCGCCAUUAUGCAUGAAUGCAUAGAAAUA ---------------------.((((..(((((((....((((.((((((((.((((....))..))))))))))))))...))))))).......((((((....)))))))))).. ( -29.90, z-score = -2.21, R) >droEre2.scaffold_4784 10020699 97 + 25762168 ---------------------UUUUCCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUA ---------------------.((((..(((((((....((((.((((((((.((((....))..))))))))))))))...))))))).......((((((....)))))))))).. ( -29.90, z-score = -2.21, R) >droAna3.scaffold_13337 14952682 99 + 23293914 -------------------UCACUUGCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUA -------------------......((.(((((((....((((.((((((((.((((....))..))))))))))))))...)))))))..))..(((((((....)))))))..... ( -30.60, z-score = -1.78, R) >dp4.chrXR_group6 5703119 101 - 13314419 -----------------UCUCGACUGCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCCGCCAUUAUGCAUGAAUGCAUAGAAAUA -----------------........((.(((((((....((((.((((((((.((((....))..))))))))))))))...)))))))..))..(((((((....)))))))..... ( -30.30, z-score = -1.79, R) >droPer1.super_27 845164 101 + 1201981 -----------------UCUCGACUACCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCCGCCAUUAUGCAUGAAUGCAUAGAAAUA -----------------...........(((((((....((((.((((((((.((((....))..))))))))))))))...)))))))......(((((((....)))))))..... ( -29.70, z-score = -2.05, R) >droWil1.scaffold_180698 7695170 96 + 11422946 ----------------------UCCACCGCAAAUGGACCGCAAAUGGCAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGACUGCAUAGAAAUA ----------------------......(((((((....((((.((((((((.((((....))..))))))))))))))...)))))))......(((((((....)))))))..... ( -31.70, z-score = -2.48, R) >droVir3.scaffold_13049 9157531 117 + 25233164 UCGCCCACGGGCCGGUUUCCCGCAGGCCGCAAAUGGACCGCAAAUGGCAGCGUGCGGAAAUCCAUGCUGCAGCUUUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAU- ..(((....)))(((....)))..(((.(((((((....(((((..((((((((........))))))))....)))))...)))))))..))).(((((((....)))))))....- ( -42.50, z-score = -1.03, R) >droGri2.scaffold_15110 4931220 114 + 24565398 ----UCUCUGGCAUGCAGGCUGCAGGCCACAAAUGGACCGCAAAUGGCAGCAUGCGGAAAUCCAUGCUGCAGCUUUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUA ----(((.((.(((((((((((..(((((....))).))(((((((((((((((........)))))))).((....))...))))))))))))....))))))....)).))).... ( -42.30, z-score = -1.75, R) >consensus _____________________UUUUCCCGCAAAUGGACCGCAAAUGGUAACGUGCGGAAAUCCAUGCUGUUGCCAUUGUAAACGUUUGCCAGCCAUUAUGCAUGAAUGCAUAGAAAUA ............................(((((((....((((.((((((((.((((....))..))))))))))))))...)))))))......(((((((....)))))))..... (-29.00 = -28.73 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:54 2011