| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,005,105 – 10,005,204 |
| Length | 99 |
| Max. P | 0.644380 |

| Location | 10,005,105 – 10,005,204 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.41472 |
| G+C content | 0.48393 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -13.68 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.644380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
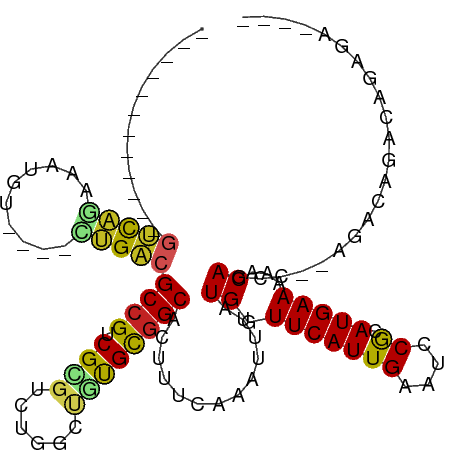
>dm3.chr3L 10005105 99 - 24543557 ------------GUCAGAAAAAU----CUGACGCCGUCGCGUCUGGCUGUGCGGCACUUUCAAAUUUAUGGUUCAUUGAAUCCGCAUGAAAAACAGCC--AGCCAGACAGACAGAGG ------------((((((....)----)))))...(((..((((((((((((((....(((((............))))).)))))((.....))..)--)))))))).)))..... ( -35.90, z-score = -3.45, R) >droSim1.chr3L 9392351 99 - 22553184 ------------GUCAGAAAAGU----CUGACGCCGUCGCGUCUGGCUGUGCGGCACUUUCAAAUUUAUGGUUCAUUGAAUCCGCAUGAAAAACAGCC--AGCCAGACAGACAGAGG ------------((((((....)----)))))...(((..((((((((((((((....(((((............))))).)))))((.....))..)--)))))))).)))..... ( -35.90, z-score = -3.16, R) >droSec1.super_0 2228284 99 - 21120651 ------------GUCAGAAAAGU----CUGACGCCGUCGUGUCUGGCUGUGCGGCACUUUCAAAUUUAUGGUUCAUUGAAUCCGCAUGAAAAACAGCC--AGCCAGACAGACAGAGG ------------((((((....)----)))))...(((.(((((((((((((((....(((((............))))).)))))((.....))..)--))))))))))))..... ( -36.90, z-score = -3.57, R) >droYak2.chr3L 9981832 95 - 24197627 ------------GUCAGAAAAGU----CUGACGCCGUCGCGUCUGGCUGUGCGGCACUUUCAAAUUUAUGGUUCAUUGAAUCCGCAUGAAAAACAGCC--AGACAGACAGAGA---- ------------((((((....)----)))))...(((..((((((((((((((....(((((............))))).)))).(....)))))))--)))).))).....---- ( -36.70, z-score = -3.90, R) >droEre2.scaffold_4784 9990444 94 - 25762168 ------------GUCAGAAAAGU----CUGACGCCGUCGCGUCUGGCUGUGCGGCACUUUCAAAUUUAUGGUUCAUUGAAUUCGCAUGAAAAACAGCC--AGACAGACGGGG----- ------------((((((....)----))))).(((((..((((((((((.((((...(((((............)))))...)).))....))))))--)))).)))))..----- ( -39.90, z-score = -4.84, R) >droAna3.scaffold_13337 14922287 91 - 23293914 ------------GUCAGCGGUCU----CUGUCGCCGUCGCGUCUGGUAGUGUGGCACUUUCAAAUUUAUGGUUCAUUGAAUCCACAUGAAAAACAGCC--AGACAGCAG-------- ------------....(((((..----.....))))).(((((((((.((((((....(((((............))))).))))))........)))--)))).))..-------- ( -25.40, z-score = -1.28, R) >dp4.chrXR_group6 5670926 99 + 13314419 ------------GUCAGAUGUCUGACGGUGACGCCGUCGAGUCUGGCUGUGCGGCACUUUCAAAUUUAUGGUUCAUUGAAUUCGCAUGAAAAACAGUC--AGACAGAUCGAGG---- ------------(((((....)))))(....).((.((((((((((((((.((((...(((((............)))))...)).))....))))))--))))...))))))---- ( -32.50, z-score = -1.63, R) >droPer1.super_27 812849 99 - 1201981 ------------GUCAGAUGUCUGACGGUGACGCCGUCGAGUCUGGCUGUGCGGCACUUUCAAAUUUAUGGUUCAUUGAAUUCGCAUGAAAAACAGUC--AGACAGAUGGAGG---- ------------(((((....)))))(....).(((((..((((((((((.((((...(((((............)))))...)).))....))))))--)))).)))))...---- ( -36.00, z-score = -2.75, R) >droWil1.scaffold_180698 7659332 97 - 11422946 -------------AUACAUAUUU---GUUGUCGCCGUCGCGUCUGGUUAUGCCGCACUUUGAAAUUUAUGGUUCAUUGAAUGCGCAUGAAAAACAAUCGGAGACAGACAAGAA---- -------------.......(((---((((((.(((....((....((((((.((((..((((........))))..)..)))))))))...))...))).)))).)))))..---- ( -22.90, z-score = -0.16, R) >droVir3.scaffold_13049 9128030 95 - 25233164 ------------GUUGUAGCUU----AGCGACGCCGUCGUGUCUGGCAGCGCGGCACUCUGAAAUUUAUGGUUCAUUGAAUGCGCAUGAAAAACAGUC--AGACAAACCGUAC---- ------------(((((.....----.)))))..((...((((((((.(((((...(..((((........))))..)..))))).((.....)))))--)))))...))...---- ( -26.10, z-score = -0.01, R) >droMoj3.scaffold_6680 18547626 110 + 24764193 GUUGUUGUUGGCAUUUCAGCUU----AGCGACGCCGUCGUGUCUGGCCACGCGGCACUUUGAAAUUUAUGGUUCAUUGAAUGCGCAUGAAAAACAGUC--AGACAAACCGAACAAC- ((((((((((((......)).)----)))(((((((.((((......)))))))).........((((((((.........)).)))))).....)))--..........))))))- ( -28.20, z-score = 0.41, R) >consensus ____________GUCAGAAAUGU____CUGACGCCGUCGCGUCUGGCUGUGCGGCACUUUCAAAUUUAUGGUUCAUUGAAUCCGCAUGAAAAACAGCC__AGACAGACAGAGA____ ............(((((..........)))))((((.((((......)))))))).............((.(((((((....)).)))))...))...................... (-13.68 = -13.60 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:53 2011