
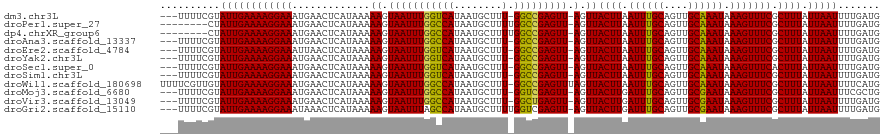
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 10,003,201 – 10,003,335 |
| Length | 134 |
| Max. P | 0.927131 |

| Location | 10,003,201 – 10,003,316 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Shannon entropy | 0.11743 |
| G+C content | 0.29269 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -21.88 |
| Energy contribution | -21.51 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 10003201 115 + 24543557 ---UUUUCGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUG ---...(((.((((((((((((.............((.(((((((((((........)-)))))))))-).))((((.((((((....)))))).))))))).)))).)))))..))).. ( -26.90, z-score = -2.56, R) >droPer1.super_27 810602 111 + 1201981 --------CUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGCCAUAAUGCUUUUGGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUG --------.(((..(((.....(((....)))..(((((((((((((((.........))))))))))-.(..((((.((((((....)))))).))))..)))))).....)))..))) ( -27.90, z-score = -2.99, R) >dp4.chrXR_group6 5668692 111 - 13314419 --------CUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGCCAUAAUGCUUUUGGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUG --------.(((..(((.....(((....)))..(((((((((((((((.........))))))))))-.(..((((.((((((....)))))).))))..)))))).....)))..))) ( -27.90, z-score = -2.99, R) >droAna3.scaffold_13337 14920469 115 + 23293914 ---UUUUCGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGCCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUG ---...(((.((((((((((((.............((.(((((((((((........)-)))))))))-).))((((.((((((....)))))).))))))).)))).)))))..))).. ( -29.60, z-score = -3.20, R) >droEre2.scaffold_4784 9988314 115 + 25762168 ---UUUUCGUAUUGAAAAGGAAAUUAACUCAUAAAAAGUAAUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUG ---...(((.((((((((((((((((((((....(((....)))(((((........)-)))))))))-))))((((.((((((....)))))).)))).)).)))).)))))..))).. ( -28.00, z-score = -3.13, R) >droYak2.chr3L 9979745 115 + 24197627 ---UUUUCGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUG ---...(((.((((((((((((.............((.(((((((((((........)-)))))))))-).))((((.((((((....)))))).))))))).)))).)))))..))).. ( -26.90, z-score = -2.56, R) >droSec1.super_0 2226380 115 + 21120651 ---UUUUCGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCACUUUAUUAAUUUUGAUG ---...(((.((((((((((((.............((.(((((((((((........)-)))))))))-).))((((.((((((....)))))).))))))).)))).)))))..))).. ( -26.90, z-score = -2.80, R) >droSim1.chr3L 9390472 115 + 22553184 ---UUUUCGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUG ---...(((.((((((((((((.............((.(((((((((((........)-)))))))))-).))((((.((((((....)))))).))))))).)))).)))))..))).. ( -26.90, z-score = -2.56, R) >droWil1.scaffold_180698 7657511 119 + 11422946 UUUUCGUUGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGCCAUAAUGCUUU-GGCCGAGUUUAGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUCAUG ((((((......))))))((((((((((((....(((....)))(((((........)-)))))))))).(..((((.((((((....)))))).))))..).........))))))... ( -31.00, z-score = -3.46, R) >droMoj3.scaffold_6680 18545334 115 - 24764193 ---UUUUCGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGCCAUAAUGCUUU-GGUCGAGUU-AGUUACUUGAUUUGCAGUUGCGAAUAAAGUUUCGCUUUAUUAAUUUCGCUG ---(((((.....))))).(((((.((((((....((((((((((((((........)-))))))...-.)))))))....)).))))(((((......))))).......))))).... ( -26.20, z-score = -1.65, R) >droVir3.scaffold_13049 9125636 115 + 25233164 ---UUUUCGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGCCAUAAUGCUUU-GGCUGAGUU-AGUUACUUGAUUUGCAGUUGCGAAUAAAGUUUCGCUUUAUUAAUUUUGAUG ---...(((.((((((((((((.............((.(((((..((((........)-)))..))))-).))((((.((((((....)))))).))))))).)))).)))))..))).. ( -26.80, z-score = -2.06, R) >droGri2.scaffold_15110 4898402 116 + 24565398 ---UUUUCGUAUUGAAAAGGAAAUAAACUCAUAAAAAGUAAUUUAGCCAUAAUGCUUUUGGUCGAGUU-AGUUACUUGAUUUGCAGUUGCGAAUAAAGUUUCGCUUUAUUAAUUUUGAUG ---...(((.((((((((((((.............((.(((((..((((.........))))..))))-).))((((.((((((....)))))).))))))).)))).)))))..))).. ( -22.10, z-score = -0.87, R) >consensus ___UUUUCGUAUUGAAAAGGAAAUGAACUCAUAAAAAGUAAUUUGGCCAUAAUGCUUU_GGCCGAGUU_AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUG .........((((((((.....(((....)))..((((((((((((((...........)))))))))..(..((((.((((((....)))))).))))..)))))).....)))))))) (-21.88 = -21.51 + -0.37)

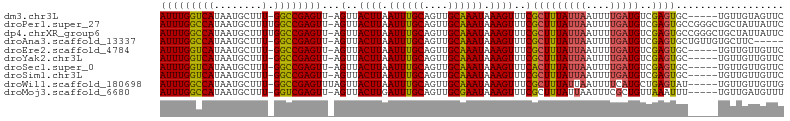
| Location | 10,003,238 – 10,003,335 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Shannon entropy | 0.21239 |
| G+C content | 0.34355 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -17.55 |
| Energy contribution | -17.34 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 10003238 97 + 24543557 AUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUGUCGAGUGC-----UGUUGUAGUUC (((((((((........)-)))))))).-...........((((((..((((((((((.....)))))))...((((....)))))))-----..))))))... ( -21.50, z-score = -1.48, R) >droPer1.super_27 810634 103 + 1201981 AUUUGGCCAUAAUGCUUUUGGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUGUCGAGUGCCGGGCUGCUAUUAUUC (((((((((.........))))))))).-.......((((..((((((((((((((((.....)))))))...((((....)))))))..)))))).))))... ( -25.80, z-score = -1.40, R) >dp4.chrXR_group6 5668724 103 - 13314419 AUUUGGCCAUAAUGCUUUUGGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUGUCGAGUGCCGGGCUGCUAUUAUUC (((((((((.........))))))))).-.......((((..((((((((((((((((.....)))))))...((((....)))))))..)))))).))))... ( -25.80, z-score = -1.40, R) >droAna3.scaffold_13337 14920506 97 + 23293914 AUUUGGCCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUGUCGAGUGCUGUUGUGCUUC----- (((((((((........)-)))))))).-.(..((((.((((((....)))))).))))..)....................(((..(....)..))).----- ( -26.20, z-score = -2.50, R) >droEre2.scaffold_4784 9988351 97 + 25762168 AUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUGUCGAGUGC-----UGUUGUUGUUC (((((((((........)-)))))))).-.(..((((.((((((....)))))).))))..)(((((((((....)))))..))))..-----........... ( -20.60, z-score = -1.25, R) >droYak2.chr3L 9979782 97 + 24197627 AUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUGUCGAGUGC-----UGUUGUUGUUC (((((((((........)-)))))))).-.(..((((.((((((....)))))).))))..)(((((((((....)))))..))))..-----........... ( -20.60, z-score = -1.25, R) >droSec1.super_0 2226417 97 + 21120651 AUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCACUUUAUUAAUUUUGAUGUCGAGUGC-----UGUUGUUGUUC (((((((((........)-)))))))).-....((((.((((((....)))))).))))..((((((((((....)))))..))))).-----........... ( -20.60, z-score = -1.35, R) >droSim1.chr3L 9390509 97 + 22553184 AUUUGGUCAUAAUGCUUU-GGCCGAGUU-AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUGUCGAGUGC-----UGUUGUUGUUC (((((((((........)-)))))))).-.(..((((.((((((....)))))).))))..)(((((((((....)))))..))))..-----........... ( -20.60, z-score = -1.25, R) >droWil1.scaffold_180698 7657551 98 + 11422946 AUUUGGCCAUAAUGCUUU-GGCCGAGUUUAGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUCAUGCUGAGUAU-----UGUUGUUGUUG (((((((((........)-))))))))...(..((((.((((((....)))))).))))..)((..((.((((.(((.....))).))-----)).))..)).. ( -23.70, z-score = -2.10, R) >droMoj3.scaffold_6680 18545371 97 - 24764193 AUUUGGCCAUAAUGCUUU-GGUCGAGUU-AGUUACUUGAUUUGCAGUUGCGAAUAAAGUUUCGCUUUAUUAAUUUCGCUGUUAAAUUU-----UGUUGAUGUUU (((((((((........)-)))))))).-.(((((..((((((((((....(((((((.....)))))))......)))).)))))).-----.).)))).... ( -22.10, z-score = -1.89, R) >consensus AUUUGGCCAUAAUGCUUU_GGCCGAGUU_AGUUACUUAAUUUGCAGUUGCAAAUAAAGUUUCGCUUUAUUAAUUUUGAUGUCGAGUGC_____UGUUGUUGUUC ((((((((...........))))))))...(..((((.((((((....)))))).))))..)(((((((((....)))))..)))).................. (-17.55 = -17.34 + -0.21)
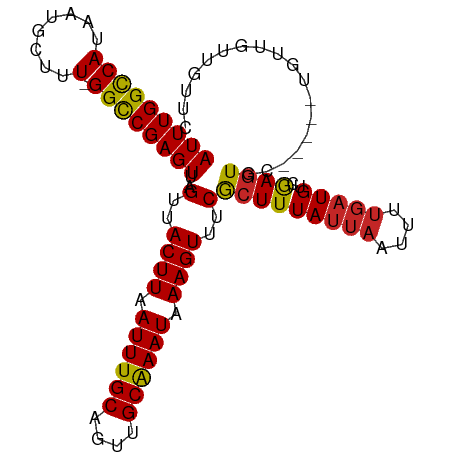
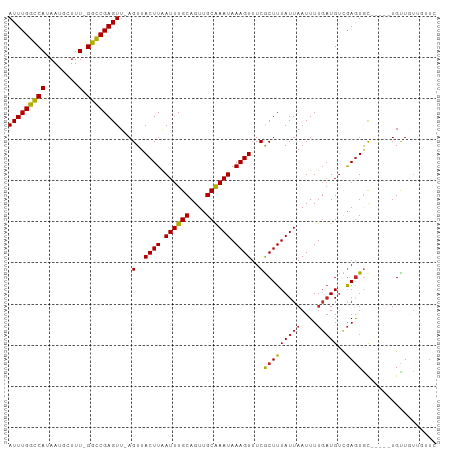
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:52 2011