
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,987,422 – 9,987,616 |
| Length | 194 |
| Max. P | 0.944041 |

| Location | 9,987,422 – 9,987,540 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Shannon entropy | 0.19465 |
| G+C content | 0.35686 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9987422 118 - 24543557 GAUGCAGCU--AGUGCAUCAUUAUCAUCUGAAAUACAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGAUAAAUUUCGGAUAACUCGCAAUUAUCGGAUAGCCAAAAUC ((((((...--..)))))).(((((..(((.....))).((((((((..(((........)))))))).)))......)))))...((.(((((.......))))).))........... ( -21.60, z-score = -0.99, R) >droEre2.scaffold_4784 9970795 118 - 25762168 GAUGCAUCU--AGUGCAUCAUUAUACCAUGAAAUACAGCACCUUUUUGGCGCACAUAAAAGUGAAAAAUGGUAAAUUAGAUUAAUUUCGCAUUACUCGCAAUUAUCUGAAAGCCAAAAUC (((((((..--.)))))))........................(((((((.((.((((..((((..(((((.((((((...))))))).))))..))))..)))).))...))))))).. ( -25.80, z-score = -2.30, R) >droYak2.chr3L 9963576 118 - 24197627 GAUGCAUCU--ACUGCAUCAUUAUUACAUGACAUACAGCACCUUUUUGGCGCACAUCAAAGUGGAAAAUGGUAAAUUAGAUUAAUUUCGAAUUACCAGCAAUUAUCUGAACGCCAAAAUC ((((((...--..))))))........................(((((((((((......))).....((((((....((......))...)))))).............)))))))).. ( -26.10, z-score = -2.57, R) >droSec1.super_0 2210695 120 - 21120651 GAUGCAUCUCCAGUGCAUCAUUAUCAUCUGAAAUUCAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUGGAUUAAAUUCGGAUUACUCGCAAUUAUCUGAUAGCCAAAAUC (((((((.....)))))))......(((((((((((((.((((((((..(((........)))))))).)))...))))))....))))))).....((.(((....))).))....... ( -23.50, z-score = -1.10, R) >droSim1.chr3L 9373223 120 - 22553184 GAUGCAUCUCUAGUGCAUCAUUAUCAUCUGAAAUGCAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGAUUAAUUUCGGAUUACUCGCAAUUAUCUGAUAGCCAAAAUC (((((((.....)))))))..................((((((((((..(((........)))))))).)))..(((((((.((((.(((.....))).))))))))))).))....... ( -24.50, z-score = -1.43, R) >consensus GAUGCAUCU__AGUGCAUCAUUAUCAUCUGAAAUACAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGAUUAAUUUCGGAUUACUCGCAAUUAUCUGAUAGCCAAAAUC (((((((.....)))))))..................((((((((((.((..........)).))))).)))...((((((.((((.(((.....))).))))))))))..))....... (-19.56 = -20.20 + 0.64)



| Location | 9,987,462 – 9,987,579 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.71 |
| Shannon entropy | 0.21291 |
| G+C content | 0.34447 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9987462 117 - 24543557 UAAAUAGGCACAG-GAAAUUAUUUUGCUAAUACGAUUAGUGAUGCAGCU--AGUGCAUCAUUAUCAUCUGAAAUACAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGA .......((..((-.(((....))).)).....(((.(((((((((...--..))))))))))))............))((((((((..(((........)))))))).)))........ ( -22.50, z-score = -0.82, R) >droEre2.scaffold_4784 9970835 113 - 25762168 UUAAUAGGAAC-----AUUUAUUUGGCUAAUACGAUUAGUGAUGCAUCU--AGUGCAUCAUUAUACCAUGAAAUACAGCACCUUUUUGGCGCACAUAAAAGUGAAAAAUGGUAAAUUAGA ...........-----..........(((((.....(((((((((((..--.)))))))))))((((((........((.((.....)).)).(((....)))....)))))).))))). ( -26.50, z-score = -2.44, R) >droYak2.chr3L 9963616 118 - 24197627 UUAAUGGGAACAGCCAAUGUAUUUGGCUAAUACGAUUAGUGAUGCAUCU--ACUGCAUCAUUAUUACAUGACAUACAGCACCUUUUUGGCGCACAUCAAAGUGGAAAAUGGUAAAUUAGA (((((......((((((.....))))))..(((.((((((((((((...--..)))))))))....(((........((.((.....)).))........)))...))).))).))))). ( -27.39, z-score = -1.30, R) >droSec1.super_0 2210735 119 - 21120651 UAAAUAGGCACAG-CAAAUUAUUUUGCUAAUACGAUUAGUGAUGCAUCUCCAGUGCAUCAUUAUCAUCUGAAAUUCAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUGGA .......((..((-((((....)))))).....(((.((((((((((.....)))))))))))))............))((((((((..(((........)))))))).)))........ ( -28.40, z-score = -2.32, R) >droSim1.chr3L 9373263 119 - 22553184 UAAAUAGGCACAG-CAAAUUAUUUUGCUAAUACGAUUAGUGAUGCAUCUCUAGUGCAUCAUUAUCAUCUGAAAUGCAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGA .......(((.((-((((....)))))).....(((.((((((((((.....)))))))))))))........)))...((((((((..(((........)))))))).)))........ ( -28.90, z-score = -2.39, R) >consensus UAAAUAGGCACAG_CAAAUUAUUUUGCUAAUACGAUUAGUGAUGCAUCU__AGUGCAUCAUUAUCAUCUGAAAUACAGCACCUUUUCAACGCACAACAAAGUGGAAAAUGGUAAAUUAGA ..........................(((((.....(((((((((((.....)))))))))))................((((((((.((..........)).))))).)))..))))). (-21.50 = -21.10 + -0.40)



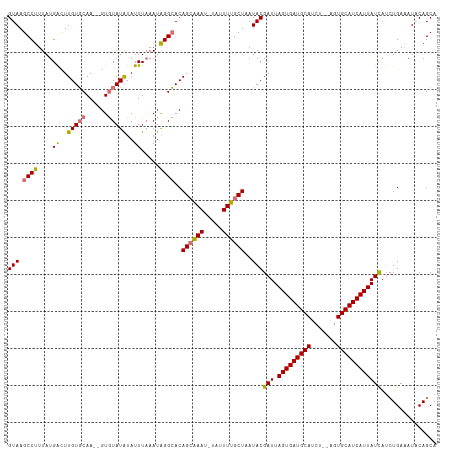
| Location | 9,987,502 – 9,987,613 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 89.20 |
| Shannon entropy | 0.17436 |
| G+C content | 0.33698 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9987502 111 - 24543557 GUAAGCCUUUAUGACUUGUGCAA--GUGUAUAUAUUUAAAUAGGCACAGGAAAU-UAUUUUGCUAAUACGAUUAGUGAUGCAGCU--AGUGCAUCAUUAUCAUCUGAAAUACAGCA (((.((((....((..(((((..--..)))))..)).....)))).((((..((-((......))))..(((.(((((((((...--..)))))))))))).))))...))).... ( -25.80, z-score = -0.71, R) >droYak2.chr3L 9963656 114 - 24197627 GUAAGCCUUUAUGGCUUGUUCAACUGUAUAUGUAUUUUAAUGGGAACAGCCAAUGUAUUUGGCUAAUACGAUUAGUGAUGCAUCU--ACUGCAUCAUUAUUACAUGACAUACAGCA .((((((.....)))))).....((((((.((((((.....(....)((((((.....))))))))))))..((((((((((...--..)))))))))).........)))))).. ( -35.00, z-score = -3.45, R) >droSec1.super_0 2210775 113 - 21120651 GUAAGCCUUUAUGAUUUGUGCAA--GUGUAUAUAUUUAAAUAGGCACAGCAAAU-UAUUUUGCUAAUACGAUUAGUGAUGCAUCUCCAGUGCAUCAUUAUCAUCUGAAAUUCAGCA (((.((((....(((.(((((..--..))))).))).....))))..((((((.-...))))))..)))(((.((((((((((.....)))))))))))))..(((.....))).. ( -32.80, z-score = -3.54, R) >droSim1.chr3L 9373303 113 - 22553184 GUAAGCCUUUAUGACUUGUGCAA--GUGUAUAUAUUUAAAUAGGCACAGCAAAU-UAUUUUGCUAAUACGAUUAGUGAUGCAUCUCUAGUGCAUCAUUAUCAUCUGAAAUGCAGCA (((.((((....((..(((((..--..)))))..)).....))))..((((((.-...))))))..)))(((.((((((((((.....)))))))))))))..(((.....))).. ( -31.00, z-score = -2.08, R) >consensus GUAAGCCUUUAUGACUUGUGCAA__GUGUAUAUAUUUAAAUAGGCACAGCAAAU_UAUUUUGCUAAUACGAUUAGUGAUGCAUCU__AGUGCAUCAUUAUCAUCUGAAAUACAGCA (((.((((....((..((((((....))))))..)).....))))..((((((.....))))))..)))(((.((((((((((.....)))))))))))))............... (-25.08 = -25.45 + 0.37)

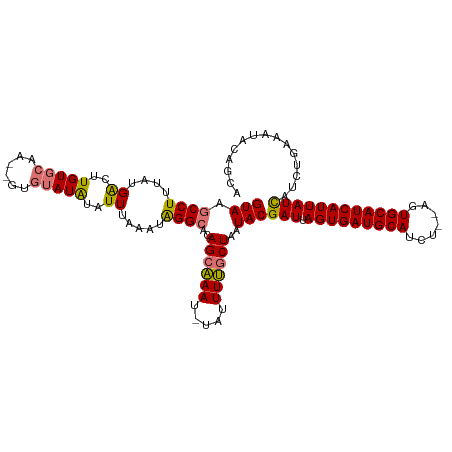
| Location | 9,987,505 – 9,987,616 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 89.20 |
| Shannon entropy | 0.17436 |
| G+C content | 0.32811 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9987505 111 - 24543557 AUGGUAAGCCUUUAUGACUUGUGCAA--GUGUAUAUAUUUAAAUAGGCACAGGAAAU-UAUUUUGCUAAUACGAUUAGUGAUGCAGCU--AGUGCAUCAUUAUCAUCUGAAAUACA .(((((((.....((..(((((((..--...((......)).....)))))))..))-...)))))))....(((.(((((((((...--..))))))))))))............ ( -26.00, z-score = -0.90, R) >droYak2.chr3L 9963659 114 - 24197627 AUGGUAAGCCUUUAUGGCUUGUUCAACUGUAUAUGUAUUUUAAUGGGAACAGCCAAUGUAUUUGGCUAAUACGAUUAGUGAUGCAUCU--ACUGCAUCAUUAUUACAUGACAUACA ....((((((.....))))))......(((.((((((...((((.(....((((((.....))))))....).))))((((((((...--..))))))))...))))))))).... ( -32.20, z-score = -2.58, R) >droSec1.super_0 2210778 113 - 21120651 AUGGUAAGCCUUUAUGAUUUGUGCAA--GUGUAUAUAUUUAAAUAGGCACAGCAAAU-UAUUUUGCUAAUACGAUUAGUGAUGCAUCUCCAGUGCAUCAUUAUCAUCUGAAAUUCA ...(((.((((....(((.(((((..--..))))).))).....))))..((((((.-...))))))..)))(((.((((((((((.....)))))))))))))............ ( -31.60, z-score = -3.07, R) >droSim1.chr3L 9373306 113 - 22553184 AUGGUAAGCCUUUAUGACUUGUGCAA--GUGUAUAUAUUUAAAUAGGCACAGCAAAU-UAUUUUGCUAAUACGAUUAGUGAUGCAUCUCUAGUGCAUCAUUAUCAUCUGAAAUGCA ...(((.((((....((..(((((..--..)))))..)).....))))..((((((.-...))))))..)))(((.((((((((((.....)))))))))))))............ ( -30.60, z-score = -2.07, R) >consensus AUGGUAAGCCUUUAUGACUUGUGCAA__GUGUAUAUAUUUAAAUAGGCACAGCAAAU_UAUUUUGCUAAUACGAUUAGUGAUGCAUCU__AGUGCAUCAUUAUCAUCUGAAAUACA ...(((.((((....((..((((((....))))))..)).....))))..((((((.....))))))..)))(((.((((((((((.....)))))))))))))............ (-25.08 = -25.45 + 0.37)

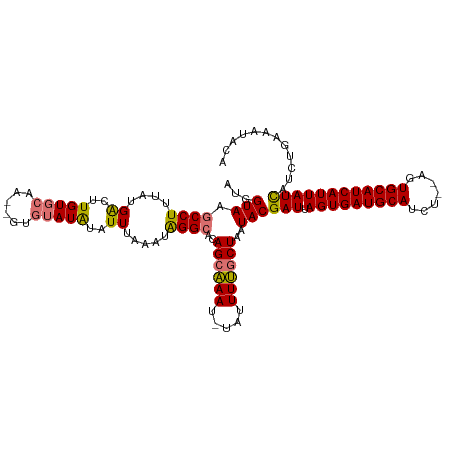

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:48 2011