| Sequence ID | dm3.chr3L |
|---|---|
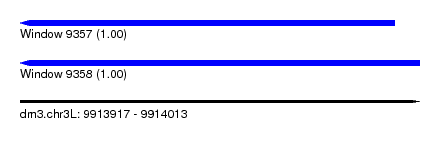
| Location | 9,913,917 – 9,914,013 |
| Length | 96 |
| Max. P | 0.997664 |

| Location | 9,913,917 – 9,914,007 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 66.21 |
| Shannon entropy | 0.61955 |
| G+C content | 0.33430 |
| Mean single sequence MFE | -14.59 |
| Consensus MFE | -7.96 |
| Energy contribution | -7.35 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997664 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
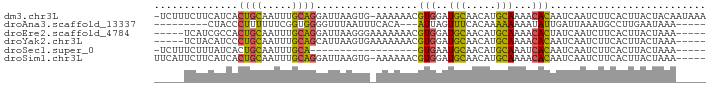
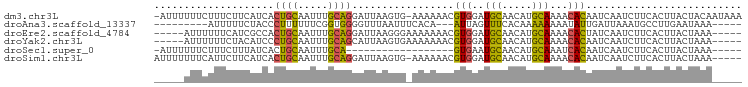
>dm3.chr3L 9913917 90 - 24543557 -UCUUUCUUCAUCACUGCAAUUUGCAGGAUUAAGUG-AAAAAACGUGGAUGCAACAUGCAAAACACAAUCAAUCUUCACUUACUACAAUAAA -.........(((.((((.....)))))))((((((-((.....(((..(((.....)))...)))........)))))))).......... ( -17.92, z-score = -3.16, R) >droAna3.scaffold_13337 14844439 75 - 23293914 ---------CUACCCUUUUUUCGGUGGGGUUUAAUUUCACA---AUUAGUUUCACAAAAAAAAUAUUGAUUAAAUGCCUUGAAUAAA----- ---------..........(((((.((.(((((((....((---((...(((......)))...))))))))))).)))))))....----- ( -9.60, z-score = 0.28, R) >droEre2.scaffold_4784 9905292 82 - 25762168 -----UCAUCGCCACUGCAAUUUGCAGGAUUAAGGGAAAAAAACGUGGAUGCAACAUGCAAAACACUAUCAAUCUUCACUUACUAAA----- -----.........((((.....))))...((((.(((......(((..(((.....)))...)))........))).)))).....----- ( -12.54, z-score = -0.75, R) >droYak2.chr3L 9890529 82 - 24197627 -----UCUACAUCCCUGCAAUUUGCAGCAUUAAGUGAAAAAAACGUGGAUGCAACAUGCAAAACACAAUCAAUCUUCACUUACUAAA----- -----.........((((.....))))...((((((((......(((..(((.....)))...)))........)))))))).....----- ( -16.14, z-score = -2.71, R) >droSec1.super_0 2146031 68 - 21120651 -UCUUUCUUUAUCACUGCAAUUUGCA------------------GUGAAUGCAACAUGCAAAUCACAAUCAAUCUUCACUUACUAAA----- -..........(((((((.....)))------------------)))).(((.....)))...........................----- ( -13.40, z-score = -3.67, R) >droSim1.chr3L 9299912 86 - 22553184 UUCAUUCUUCAUCACUGCAAUUUGCAGGAUUAAGUG-AAAAAACGUGGAUGCAACAUGCAAAACACAAUCAAUCUUCACUUACUAAA----- ..........(((.((((.....)))))))((((((-((.....(((..(((.....)))...)))........)))))))).....----- ( -17.92, z-score = -2.93, R) >consensus _____UCUUCAUCACUGCAAUUUGCAGGAUUAAGUG_AAAAAACGUGGAUGCAACAUGCAAAACACAAUCAAUCUUCACUUACUAAA_____ ..............((((.....)))).................(((..(((.....)))...))).......................... ( -7.96 = -7.35 + -0.61)
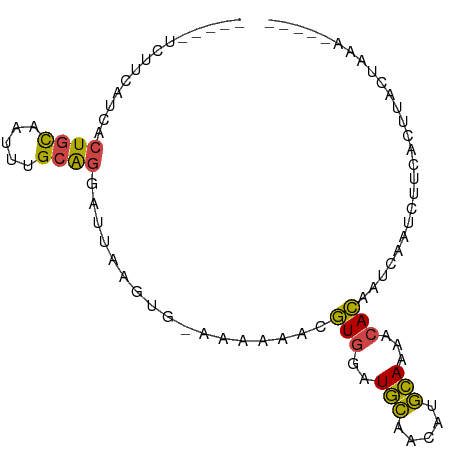
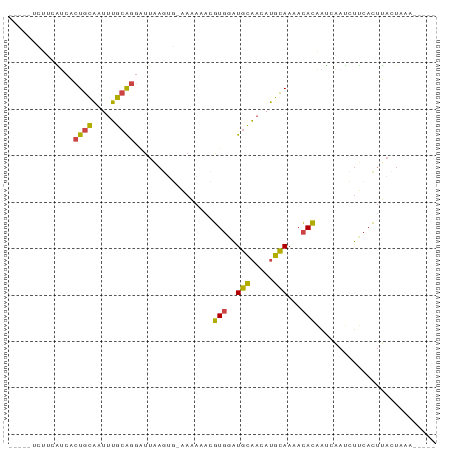
| Location | 9,913,917 – 9,914,013 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 66.45 |
| Shannon entropy | 0.61512 |
| G+C content | 0.31102 |
| Mean single sequence MFE | -15.85 |
| Consensus MFE | -7.96 |
| Energy contribution | -7.35 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.996806 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
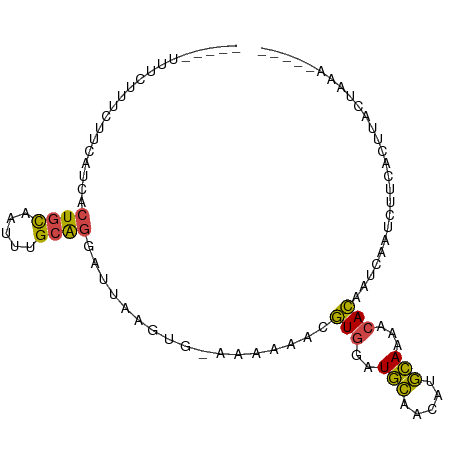
>dm3.chr3L 9913917 96 - 24543557 -AUUUUUUCUUUCUUCAUCACUGCAAUUUGCAGGAUUAAGUG-AAAAAACGUGGAUGCAACAUGCAAAACACAAUCAAUCUUCACUUACUACAAUAAA -.(((((((...(((.(((.((((.....))))))).))).)-)))))).(((..(((.....)))...))).......................... ( -20.10, z-score = -3.54, R) >droAna3.scaffold_13337 14844439 81 - 23293914 ---------AUUUUUCUACCCUUUUUUCGGUGGGGUUUAAUUUCACA---AUUAGUUUCACAAAAAAAAUAUUGAUUAAAUGCCUUGAAUAAA----- ---------................(((((.((.(((((((....((---((...(((......)))...))))))))))).)))))))....----- ( -9.60, z-score = 0.45, R) >droEre2.scaffold_4784 9905292 88 - 25762168 -----AUUUUUUCAUCGCCACUGCAAUUUGCAGGAUUAAGGGAAAAAAACGUGGAUGCAACAUGCAAAACACUAUCAAUCUUCACUUACUAAA----- -----.(((((((.......((((.....)))).......)))))))...(((..(((.....)))...))).....................----- ( -15.74, z-score = -1.71, R) >droYak2.chr3L 9890529 88 - 24197627 -----AUUUUUUCUACAUCCCUGCAAUUUGCAGCAUUAAGUGAAAAAAACGUGGAUGCAACAUGCAAAACACAAUCAAUCUUCACUUACUAAA----- -----...............((((.....))))...((((((((......(((..(((.....)))...)))........)))))))).....----- ( -16.14, z-score = -2.28, R) >droSec1.super_0 2146031 74 - 21120651 -AUUUUUUCUUUCUUUAUCACUGCAAUUUGCA------------------GUGAAUGCAACAUGCAAAUCACAAUCAAUCUUCACUUACUAAA----- -................(((((((.....)))------------------)))).(((.....)))...........................----- ( -13.40, z-score = -3.16, R) >droSim1.chr3L 9299912 92 - 22553184 AUUUUUUUCAUUCUUCAUCACUGCAAUUUGCAGGAUUAAGUG-AAAAAACGUGGAUGCAACAUGCAAAACACAAUCAAUCUUCACUUACUAAA----- ..((((((((..(((.(((.((((.....))))))).)))))-)))))).(((..(((.....)))...))).....................----- ( -20.10, z-score = -3.45, R) >consensus _____UUUCUUUCUUCAUCACUGCAAUUUGCAGGAUUAAGUG_AAAAAACGUGGAUGCAACAUGCAAAACACAAUCAAUCUUCACUUACUAAA_____ ....................((((.....)))).................(((..(((.....)))...))).......................... ( -7.96 = -7.35 + -0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:42 2011