
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,898,354 – 9,898,451 |
| Length | 97 |
| Max. P | 0.809295 |

| Location | 9,898,354 – 9,898,451 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.88 |
| Shannon entropy | 0.52203 |
| G+C content | 0.51176 |
| Mean single sequence MFE | -22.01 |
| Consensus MFE | -11.07 |
| Energy contribution | -11.37 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9898354 97 - 24543557 UGUUAGCCCAUCCUCUCCGCCCAUAGACGAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGCUCAAACAAAGCAAGGACCA--AACCUGUAAACGGCCC---------- .....(((.........((((((..((((..((((((......)))))))))).)).))))..........(((.((....--..)))))....)))..---------- ( -22.60, z-score = -1.65, R) >droSim1.chr3L 9284095 97 - 22553184 UGUUGGCCCAUCCUCCCCGCCCAUAGACAAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGCUCAAACAAAGCAAGGACCA--AACCUGUAAACGGCCC---------- ....((((.........((((((..(((...((((((......)))))).))).)).))))..........(((.((....--..)))))....)))).---------- ( -23.40, z-score = -1.51, R) >droSec1.super_0 2130628 97 - 21120651 UGUUGGCCCAUCCUCCCCGCCCAUAGACAAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGCUCAAACAAAGCAAGGACCA--AACCUGUAAACGGCCC---------- ....((((.........((((((..(((...((((((......)))))).))).)).))))..........(((.((....--..)))))....)))).---------- ( -23.40, z-score = -1.51, R) >droYak2.chr3L 9874791 83 - 24197627 CGUUGGCCCAUCCU--CCGCCCAUAGACAAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGCUCAAACAAAGCAAGGACCA--AACC---------------------- ..((((.((.....--..(((((..(((...((((((......)))))).))).)).)))(((.......)))..)).)))--)...---------------------- ( -21.40, z-score = -2.38, R) >droEre2.scaffold_4784 9890041 83 - 25762168 UGCUGGCCCCUCCU--CCGCCCAUAGACAAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGCGCAAACAAAGCAAGGACCA--AGCC---------------------- .(((((((......--.((((((..(((...((((((......)))))).))).)).)))).((.......))..)).)).--))).---------------------- ( -21.90, z-score = -1.37, R) >droAna3.scaffold_13337 14827818 95 - 23293914 UAUUUGCCCAUCCU--CCGCACAUAGACAAACUCAUUACGAGAAAUGAGCGUCCUGCGGCGCUCAAACAAAGCAAGGACCA--AACCUGUAAACGGCCC---------- .....(((......--..((...........(((.....)))...((((((((....))))))))......)).(((....--..)))......)))..---------- ( -22.60, z-score = -1.96, R) >dp4.chrXR_group6 5566989 95 + 13314419 UAUUUGCUCAUCCU--CUGCAGAUAGACAAACUCAUUACUAAAAAUGAGCCUCCUGCGGCGUUCAAACAAAGCAAGGACCA--AACCUGUAAACGGGCC---------- (((((((.......--..)))))))......((((((......))))))..((((...((...........)).))))...--..((((....))))..---------- ( -20.00, z-score = -1.04, R) >droPer1.super_27 708635 95 - 1201981 UAUUUGCUCAUCCU--CUGCAGAUAGACAAACUCAUUACUAAAAAUGAGCCUCCUGCGGCGUUCAAACAAAGCAAGGACCA--AACCUGUAAACGGGCC---------- (((((((.......--..)))))))......((((((......))))))..((((...((...........)).))))...--..((((....))))..---------- ( -20.00, z-score = -1.04, R) >droWil1.scaffold_180698 5006869 76 + 11422946 ---------------------CAUAGACAAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGUUCAAACAAAGCAGGGACCA--UACCUGAGAGACCCAC---------- ---------------------..........(((...........((((((((....)))))))).......((((.....--..)))).)))......---------- ( -18.60, z-score = -2.58, R) >droVir3.scaffold_13049 9027261 94 - 25233164 ----UGCUCCACC-UGAACA---CGGCCAAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGUUCAAACAAAGCAAAGUCCCCGUACCUGAGGGCGGCCAACA------- ----((((.....-(((((.---(.((....((((((......))))))......)).).))))).....))))..((((((........))).))).....------- ( -24.70, z-score = -0.63, R) >droGri2.scaffold_15110 4802187 103 - 24565398 -----GCUUCACCUUGCACAUACUAGGCAAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGUUCAAACAAAGCAAGGUUCUC-CACCUGAGGGCAUACAACACCAGACC -----(((..(((((((.......((.....))............((((((((....))))))))......))))))).(((-.....))))))............... ( -23.50, z-score = -1.08, R) >consensus UGUUGGCCCAUCCU__CCGCCCAUAGACAAACUCAUUACGAAAAAUGAGCGUCCUGCGGCGCUCAAACAAAGCAAGGACCA__AACCUGUAAACGGCCC__________ ...............................((((((......)))))).(((((...((...........)).))))).............................. (-11.07 = -11.37 + 0.31)

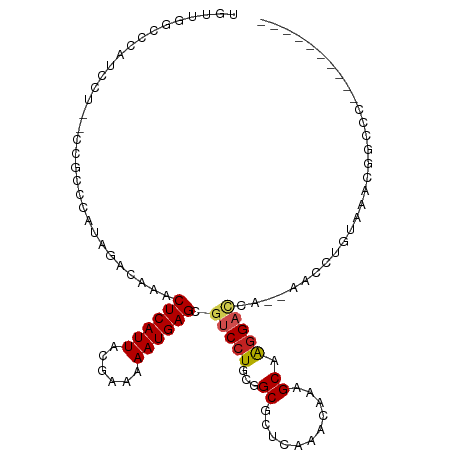
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:38 2011