| Sequence ID | dm3.chr2L |
|---|---|
| Location | 519,304 – 519,396 |
| Length | 92 |
| Max. P | 0.985596 |

| Location | 519,304 – 519,396 |
|---|---|
| Length | 92 |
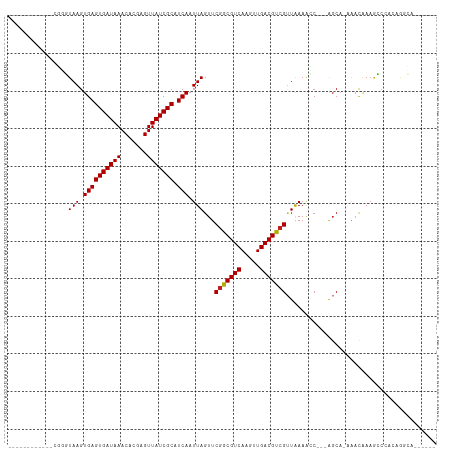
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.15 |
| Shannon entropy | 0.45086 |
| G+C content | 0.47227 |
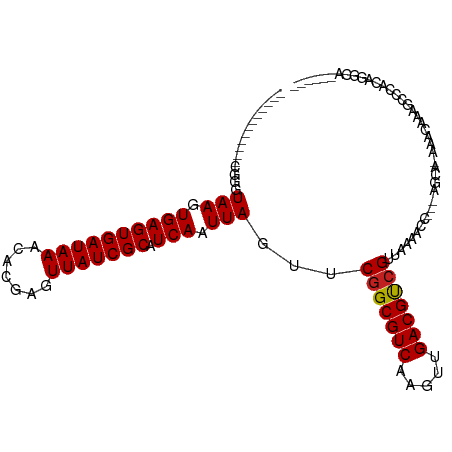
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 519304 92 - 23011544 ------------CGGGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAACC---AGCA-AAACAAAGCCCACAGGCA------ ------------..(((...((((((((((.......))))))).))).......(((((((.....))))))).....)))---....-.......(((....))).------ ( -24.70, z-score = -2.46, R) >droWil1.scaffold_180772 6084960 96 + 8906247 ------------UGGUUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGCAGCUGAAAACAAAGCCUCGAACGAAGACGAAAGUCA------ ------------....(((.((((((((((.......))))))).))).)))((((((((((.....))))).(((........)))..)))))...(((....))).------ ( -31.10, z-score = -3.34, R) >droPer1.super_1 7721635 108 + 10282868 CGCUCUCGCUCCCUUCUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAGGCAGCAGCAGAAACAAAAGCCACGAAGC------ .(((.(((((.((((((((.((((((((((.......))))))).))).))))..(((((((.....)))))))...)))).))).((..........))...)))))------ ( -30.50, z-score = -1.86, R) >dp4.chr4_group3 10608620 108 + 11692001 CGCUCUCGCUCCCUUCUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAGGCAGCAGCAGAAACAAAAGCCACGAAGC------ .(((.(((((.((((((((.((((((((((.......))))))).))).))))..(((((((.....)))))))...)))).))).((..........))...)))))------ ( -30.50, z-score = -1.86, R) >droAna3.scaffold_12943 328395 100 + 5039921 ------------UGGGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAGGCA-GAGCC-AAACAAAGCCCCGGGGCCGGGCCA ------------.((.(((.((((((((((.......))))))).))).)))(((.((((((.....)))(((......))).-..)))-.))).....((((....)))))). ( -29.80, z-score = -1.05, R) >droEre2.scaffold_4929 574166 92 - 26641161 ------------UGGGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAACC---AGCA-AAACAAAGCCCCCAGGCA------ ------------(((.(((.((((((((((.......))))))).))).)))...(((((((.....)))))))......))---)...-.......(((....))).------ ( -25.00, z-score = -2.42, R) >droYak2.chr2L 505855 92 - 22324452 ------------CGGGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAACC---AGCA-AAACAAAGCCCCCAGGCA------ ------------..(((...((((((((((.......))))))).))).......(((((((.....))))))).....)))---....-.......(((....))).------ ( -24.70, z-score = -2.38, R) >droSec1.super_14 501936 92 - 2068291 ------------CGGGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAACC---AGCA-AAACAAAGCCCGCAGGCA------ ------------(((((...((((((((((.......))))))).))).......(((((((.....)))))))........---....-.......)))))......------ ( -25.40, z-score = -2.25, R) >droSim1.chr2L 527522 92 - 22036055 ------------CGGGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAACC---AGCA-AAACAAAGCCCGCAGGCA------ ------------(((((...((((((((((.......))))))).))).......(((((((.....)))))))........---....-.......)))))......------ ( -25.40, z-score = -2.25, R) >droMoj3.scaffold_6500 8125483 86 + 32352404 ----CUGUCUGU-CCGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAACUUGACGUCG-UAAAGCC---AGCA-CGGUAA------------------ ----(((((((.-...(((.((((((((((.......))))))).))).)))...(((((((.....)))))))-......)---)).)-)))...------------------ ( -23.50, z-score = -1.45, R) >droGri2.scaffold_15252 6862161 87 - 17193109 --------CCGC-CCGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAACUUGACGUCGUUAAAGGC---AGCA-AAACAAAGCC-------------- --------..((-(..(((.((((((((((.......))))))).))).......(((((((.....))))))))))..)))---.((.-.......)).-------------- ( -24.00, z-score = -2.47, R) >droVir3.scaffold_12963 18955547 103 - 20206255 UCUCCCGUCUCUGCCGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAACUUGACGUCGUUAAAGGC---AGCA-CACCAAAGCCUGCAAGC------- ......((..(((((.(((.((((((((((.......))))))).))).)))...(((((((.....))))))).....)))---)).)-)................------- ( -27.50, z-score = -1.46, R) >consensus ____________CGGGUAAGUGAGUGAUAAACACGAGUUAUCGCAUCAAUUAGUUCGGCGUCAAGUUGACGUCGUUAAAACC___AGCA_AAACAAAGCCCACAGGCA______ ................(((.((((((((((.......))))))).))).)))...(((((((.....)))))))........................................ (-17.76 = -17.77 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:03 2011