| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,894,962 – 9,895,052 |
| Length | 90 |
| Max. P | 0.768047 |

| Location | 9,894,962 – 9,895,052 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Shannon entropy | 0.35642 |
| G+C content | 0.27237 |
| Mean single sequence MFE | -13.52 |
| Consensus MFE | -8.91 |
| Energy contribution | -9.16 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768047 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
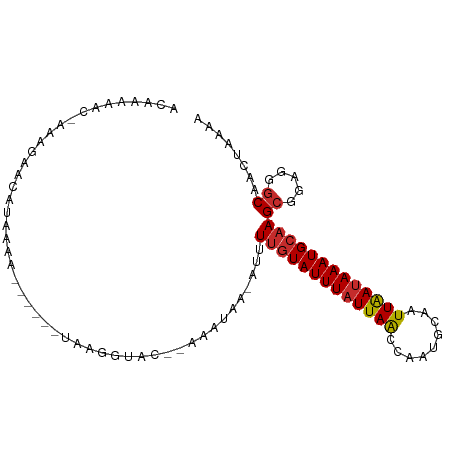
>dm3.chr3L 9894962 90 - 24543557 ACAAAAAC-AAAGAACAUAAAA------UAAGGUAC--AAAUAA-AUUUUGUAUUUAUUAACCAAUGCAAUUAAUAAAUGCAAGCGGAGGGCAACUAAAA ........-.............------........--......-..((((((((((((((.........))))))))))))))....((....)).... ( -11.30, z-score = -1.24, R) >droSim1.chr3L 9280746 90 - 22553184 ACAAAAAC-AAAGAACAUAAAA------UAAGGUAC--AAAUAA-GUUUUGUAUUUAUUAACCAAUGCAAUUAAUAAAUGCAAGCGGAGGGCAACUAAAA ........-.............------........--......-((((.(((((((((((.........)))))))))))))))...((....)).... ( -11.70, z-score = -1.03, R) >droSec1.super_0 2127365 90 - 21120651 ACAAAAAC-AAAGAACAUAAAA------UAAGGUAC--AAAUAA-GUUUUGUAUUUAUUAACCAAUGCAAUUAAUAAAUGCAAGCGGAGGGCAACUAAAA ........-.............------........--......-((((.(((((((((((.........)))))))))))))))...((....)).... ( -11.70, z-score = -1.03, R) >droYak2.chr3L 9871287 90 - 24197627 ACAAAAAC-AAAGAACAUAAAA------UAAGGUAC--AAAUAA-GUUUUGUAUUUAUUAACCAAUGCAAUUAAUAAAUGCAAGCGGAGGGCAACUAAAA ........-.............------........--......-((((.(((((((((((.........)))))))))))))))...((....)).... ( -11.70, z-score = -1.03, R) >droEre2.scaffold_4784 9886590 90 - 25762168 ACAAAAAC-AAAGAACAUAAAA------UAAGGUAC--AAAUAA-GUAUUGUAUUUAUUAACCAAUGCAAUUAAUAAAUGCAAGCGGAGGGCAACUAAAA ........-.............------........--......-((.(((((((((((((.........)))))))))))))))...((....)).... ( -11.70, z-score = -1.27, R) >droAna3.scaffold_13337 14824798 91 - 23293914 ACAAAAACAAAAGACCGCAAAA------UAAGGUAC--AAAUAA-AAUUUGUAUUUAUUAGCCAAUGCAAUUGACAAAUGCAAGCGGUGAGCAAUUAAAG .............(((((....------..((((((--((((..-.)))))))))).........((((.((....)))))).)))))............ ( -16.70, z-score = -2.17, R) >dp4.chrXR_group6 5563581 98 + 13314419 ACAGUAACAACAAAACAAAAGAUGUCCGUAAGGUAA--AAAAAAUAGUUUGUAUUUAUUAACCAAAGCAAUUAAUAAAUGCAAGCGUCGAGCACUUAAAA ..................(((.(((((....))...--........(((((((((((((((.........))))))))))))))).....)))))).... ( -18.00, z-score = -2.97, R) >droPer1.super_27 704411 98 - 1201981 ACAGUAACAACAAAACAAAAGAUGUCCGUAAGGUAA--AAAAAAUAGUUUGUAUUUAUUAACCAAAGCAAUUAAUAAAUGCAAGCGUCGAGCACUUAAAA ..................(((.(((((....))...--........(((((((((((((((.........))))))))))))))).....)))))).... ( -18.00, z-score = -2.97, R) >droVir3.scaffold_13049 9023917 96 - 25233164 ACGGCAACAAAAAACAAACAUAGAU---UAAGUUACAAAAAUAAAAAUUGUUAUUUAUUAACAAAAGCAAUUAAUAAAUGCAAGCGCCAAACAAAGACA- ..(((....................---...................((((((.....))))))..(((.........)))....)))...........- ( -10.90, z-score = -1.54, R) >consensus ACAAAAAC_AAAGAACAUAAAA______UAAGGUAC__AAAUAA_AUUUUGUAUUUAUUAACCAAUGCAAUUAAUAAAUGCAAGCGGAGGGCAACUAAAA ................................................(((((((((((((.........)))))))))))))((.....))........ ( -8.91 = -9.16 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:37 2011