
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,893,658 – 9,893,798 |
| Length | 140 |
| Max. P | 0.723714 |

| Location | 9,893,658 – 9,893,798 |
|---|---|
| Length | 140 |
| Sequences | 14 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.50277 |
| G+C content | 0.46658 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -12.02 |
| Energy contribution | -11.81 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.93 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9893658 140 - 24543557 ACCUCUCGUCUCAUCCAUUAAUCGCUGUAGUUUUUCCUGCAGCAAUCCCUUUUCAUCCACUUCCAGCUCCAGAAGGGCGUUCUUCUCGUAGGCCUGAUCCAGCAUCUUCUCGAAAUUCUCUAUGCUUUCGUUAAGUAUGC ...((..(((((...........(((((((......)))))))...(((((((.................)))))))..........).))))..))....(((((((..(((((..........)))))..))).)))) ( -27.93, z-score = -1.81, R) >droSim1.chr3L 9279444 140 - 22553184 ACCUCUCGUCUCAUCCAUUAAUCGCUGUAGUUUCUCCUGCAGCAAACCCUUUUCAUCCACUUCCAGCUCCAGGAGGGCGUUCUUCUCGUAGGCCUGAUCCAGCAUCUUCUCGAAAUUUUCUAUGCUUUCGUUAAGUAUAC .....(((...............(((((((......)))))))......................(((.((((...(((.......)))...))))....))).......))).......(((((((.....))))))). ( -27.20, z-score = -1.40, R) >droSec1.super_0 2126071 140 - 21120651 ACCUCUCGUCUCAUCCAUUAAUCGCUGUAGUUUUUCCUGCAGCAAACCCUUUUCAUCCACUUCCAGCUCCAGGAGGGCGUUCUUCUCGUAGGCCUGAUCCAGCAUCUUCUCGAAAUUCUCUAUGCUUUCGUUAAGUAUAC .....(((...............(((((((......)))))))......................(((.((((...(((.......)))...))))....))).......))).......(((((((.....))))))). ( -27.20, z-score = -1.40, R) >droYak2.chr3L 9869998 140 - 24197627 ACCUCUCGUCUCAUCCAUCAACCGCUGCAGUUUUUCCUGCAGCAAUCCCUUUUCAUCCACUUCCAGCUCUAGAAGGGCGUUCUUCUCGUAGGCCUGAUCCAGCAUUUUCUCGAAAUUCUCUAUGCUUUCGUUGAGUAUGC ...((..(((((...........(((((((......)))))))......................((((.....)))).........).))))..))....((((.(((.(((((..........)))))..))).)))) ( -29.20, z-score = -1.27, R) >droEre2.scaffold_4784 9885297 140 - 25762168 ACCUCUCGUCUCAUCCAUCAACCGCUGCAGUUUUUCCUGCAGCAAUCCCUUUUCAUCCACUUCCAGCUCUAGAAGGGCGUUCUUUUCGUAGGCCUGAUCCAGCAUUUUCUCGAAAUUCUCUAUGCUUUCAUUGAGUAUGC ......(((((((..........(((((((......)))))))....................(((..(((((((((....)))))).)))..)))....(((((................))))).....)))).))). ( -28.19, z-score = -1.23, R) >droAna3.scaffold_13337 14823416 139 - 23293914 -CCUCUCGUCUCAUCCAUCAACCGCUGCAGCUUCUCCUGCAACACACCCUUUUCGUCUACCUCCAGCUCGAGAAGAGCGUUCUUCUCGUACGCCACAUCCAACAUCUUCUCGAAAUUCUCUAUGCUUUCGUUGAGUAUGC -..........(((...(((((.(.(((((......))))).)........................(((((((((((((.(.....).))))...........)))))))))................)))))..))). ( -25.80, z-score = -1.91, R) >dp4.chrXR_group6 5562147 140 + 13314419 ACCUCGCGUCUCAUCCAUUAGCCGUUGCAGCUUCUCUUGCAGCAGACCCUUUUCAUCCACCUCCAACUCGAGCAGAGCAUUCUUCUCAUAUGCUUGAUCCAACAUCUUUUCAAAGUUCUCUAUGCUUUCGUUGAGUAUGC .....((((((((......(((.(((((((......)))))))(((..((((...............((((((((((.......)))...)))))))..............))))...)))..))).....)))).)))) ( -26.85, z-score = -1.24, R) >droPer1.super_27 702977 140 - 1201981 ACCUCGCGUCUCAUCCAUUAGCCGUUGCAGCUUCUCUUGCAGCAGACCCUUUUCAUCCACCUCCAACUCGAGCAGAGCAUUCUUCUCAUAUGCUUGAUCCAACAUCUUUUCAAAGUUCUCUAUGCUUUCGUUGAGUAUGC .....((((((((......(((.(((((((......)))))))(((..((((...............((((((((((.......)))...)))))))..............))))...)))..))).....)))).)))) ( -26.85, z-score = -1.24, R) >droWil1.scaffold_180698 4999920 140 + 11422946 ACCUCGCGUCUCAUCCAUGAGCCGCUGCAGUUUUUCCUGCAGCAAUCCUUUCUCGUCUACCUCCAGUUCAAGCAGGGCAUUCUUUUCGUAUGCCUGAUCCAACAUUUUCUCAAAAUUCUCUAUGCUCUCGUUGAGAAUGC .....((((((((.....((((.(((((((......)))))))......................(((......((((((.(.....).)))))).....)))....................))))....)))).)))) ( -34.70, z-score = -3.10, R) >droVir3.scaffold_13049 9022484 140 - 25233164 ACCGCGGGUCUCGUCCAUUAGCCGCUGCAGCUUCUCCUGCAGCAUUUCCUUUUCGUCCACCUCCAUCUCGAGAAGUGCAUUUUUCUCAUAGGCUUUGUCCAACAUUUUCUCAAAGUCUACUAUGCUCUCAUUGAGUAUGC ...(.((((..((..........(((((((......)))))))..........))...)))).).....((((((......)))))).(((((((((.............))))))))).(((((((.....))))))). ( -37.67, z-score = -3.83, R) >droMoj3.scaffold_6680 18444653 140 + 24764193 ACCACGAGUCUCGUCCAUUAGUCGCUGUAACUUCUCUUGCAGAACCUCCUUCUCGUCCACCUCCAUCUCAAGGAGUGCAUUCUUUUCAUAGGCUUUGUCCAACAUCUUCUCAAAGUCUACUAUGCUCUCGUUGAGUAUGC ...(((((...((.(.....).))((((((......)))))).........))))).......((((((((.(((.((((........(((((((((.............)))))))))..)))).))).))))).))). ( -33.12, z-score = -3.28, R) >droGri2.scaffold_15110 4796307 140 - 24565398 ACCGCGGGUCUCGUCCAUUAGCCGCUGCAGCUUCUCUUGCAGCAUCUCCUUCUCGUCCACCUCCAUCUCAAGGAGCGCAUUCUUUUCAUAGGCUUUGUCCAACAUCUUCUCAAAGUCUACUAUGCUCUCGUUGAGUAUUC ...(.((((..((......((..(((((((......)))))))..))......))...)))).)..(((((.(((.((((........(((((((((.............)))))))))..)))).))).)))))..... ( -37.22, z-score = -4.06, R) >anoGam1.chr3R 7372569 140 + 53272125 AUCGCGCGCCUCAUCCAUUAAUCGCUGCAGCUUAAUCUGCAUGCGCUCCUUCUCGUCCACCUCCAGCUCGAGCAGUGCGUUCUUCUCGUACGCCUGGUUCAGCAUCGAUUCGAAUUCGGCCACACUUUCGCUCGCUACCC ...(((.((.............((((((((......))))).))).........(.((.......(((.((.(((.((((.(.....).))))))).)).))).(((...)))....)).)........)).)))..... ( -33.00, z-score = -0.54, R) >apiMel3.GroupUn 43091088 140 - 399230636 ACCUCUUAUUUCAUCCAUUAGUCUUUGUACUAUAACUUUUAAAGCUUCUUUUUCAUCAAGUUCUGAUUCUAACAAAGCAUUCUUUUCAAUAGCAGAAUUAAGUUUAGCUUCUAUUUCUUCUUCUGUUACCCUAUUUAUUC ..................((((......))))...........((((..((...((((.....))))...))..))))...........((((((((..(((..(((...)))...))).))))))))............ ( -13.10, z-score = -0.52, R) >consensus ACCUCGCGUCUCAUCCAUUAACCGCUGCAGCUUCUCCUGCAGCAAUCCCUUUUCAUCCACCUCCAGCUCGAGAAGGGCAUUCUUCUCGUAGGCCUGAUCCAACAUCUUCUCGAAAUUCUCUAUGCUUUCGUUGAGUAUGC .......................(((((((......))))))).............................................................................(((((((.....))))))). (-12.02 = -11.81 + -0.21)

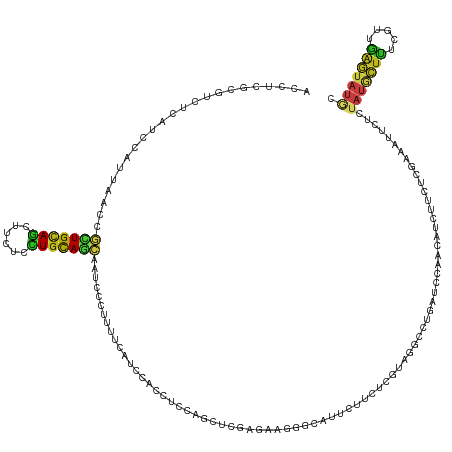
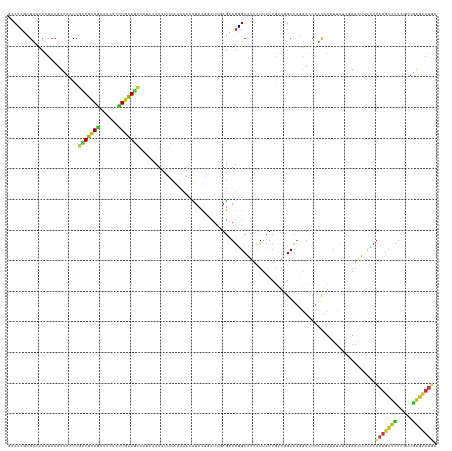
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:36 2011