| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,879,148 – 9,879,240 |
| Length | 92 |
| Max. P | 0.826666 |

| Location | 9,879,148 – 9,879,240 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 64.42 |
| Shannon entropy | 0.75162 |
| G+C content | 0.41853 |
| Mean single sequence MFE | -19.75 |
| Consensus MFE | -12.72 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.04 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
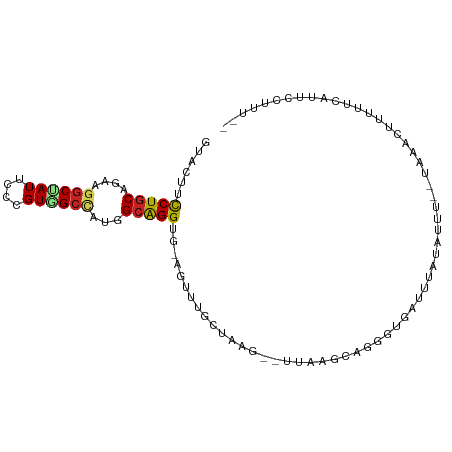
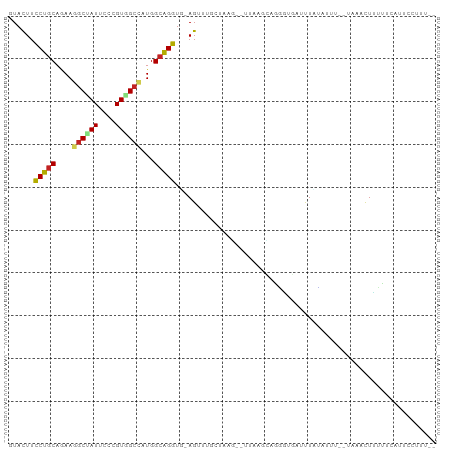
>dm3.chr3L 9879148 92 - 24543557 GUACUUCCUGCAGAAGGCUAUUCCCGUAGCCAUGGCAGGUU-AGUUUGCUUAG--CUAAGCAGGGUGAUUUAU-UUU--AAAACUUUUGCAUUCCUUU-- ........(((((((((((((....)))))).....(((((-(.(((((((..--..))))))).))))))..-...--.....))))))).......-- ( -24.50, z-score = -0.91, R) >droSim1.chr3L 9262789 92 - 22553184 GUACUUCCUGCAGAAGGCUAUUCCCGUGGCCAUGGCAGGUU-AGUUUGCUUAG--CUAAGCAGGGUGAUUUAU-UUG--AAAACUUUUGCAUUCCUUU-- ........(((((((((((((..(....)..)))))(((((-(.(((((((..--..))))))).))))))..-...--....)))))))).......-- ( -24.80, z-score = -0.38, R) >droSec1.super_0 2111492 92 - 21120651 GUACUUCCUGCAGAAGGCUAUUCCCGUGGCCAUGGCAGGUU-AGAUUGCUUAG--CCAAGUAGGGUGAUUUAU-UUU--AAAACUUUUGCAUUCCUUU-- ...(.((((((....((((((....)))))).((((((((.-.....)))).)--))).)))))).)......-...--...................-- ( -24.10, z-score = -0.61, R) >droYak2.chr3L 9855396 93 - 24197627 GUACUUCCUGCAGAAAGCUAUUCCCGUGGCCAUGGCAGGUC-AGUUGGUUGAG--UUAAGCUUGGUGAUUUCUUCUA--UAAACUUUUGCUUUAUUUU-- ..(((.(((((.....(((((....)))))....)))))..-))).....(((--(.((((..(((...........--...)))...)))).)))).-- ( -18.04, z-score = 0.85, R) >droEre2.scaffold_4784 9869382 93 - 25762168 GUACUUCCUGCAGAAGGCUAUUCCCGUGGCCAUGGCAGGUC-AGUUGUUUGCG--UUUAACCGGGUGAUUUAUAUUA--AAAACUAUUGCAUUCUUUU-- ..(((.(((((....((((((....))))))...)))))..-))).(..((((--..........((((....))))--........))))..)....-- ( -19.57, z-score = 0.36, R) >droAna3.scaffold_13337 14809261 98 - 23293914 CUAUUUCCUGCAGAAAGCGAUCCCUGUGGCCAUGGCAGGUAGAGUAUUCCAUUAAUCCUACUAUAUUAUCUUAAUCUAAUCACUUUUUUAAUUUCUUU-- .((((((((((.....((.((....)).))....)))))..)))))....................................................-- ( -10.80, z-score = 1.30, R) >dp4.chrXR_group6 5548022 84 + 13314419 GUACUUUCUGCAAAAGGCGAUUCCUGUGGCUAUGGCAGGUG-AG------------UAUUGAGUCCUUCUUUGAUCC--UUAACUGAUUCGGUUCCCUG- (((((((((((....(((.((....)).)))...))))).)-))------------)))((((((............--......))))))........- ( -16.87, z-score = 0.59, R) >droPer1.super_27 688892 84 - 1201981 GUACUUUCUGCAAAAGGCGAUUCCUGUGGCUAUGGCAGGUG-AG------------UAUUGAGUCCUUCUUUGAUCC--UUAACUGAUUCGGUUCUCUG- ......(((((....(((.((....)).)))...))))).(-((------------.((((((((............--......)))))))).)))..- ( -18.57, z-score = -0.07, R) >droWil1.scaffold_180698 4984879 96 + 11422946 AUACUUUCUGCAAAAGUCCAUUCCAGUGGCUAUGGCAGGUA-AGU-UUGGAUC--UCCAAGUGAACUAGCUAUAUCUUAUUAACUUUUUAUCUCUUAUAA ...........((((((((((....)))).((((((.(((.-.((-((((...--.))))))..))).))))))........))))))............ ( -17.20, z-score = -0.32, R) >droVir3.scaffold_13049 9007854 93 - 25233164 GUACUUCCUGCAAAAGGCCAUUCCCGUGGCCAUGGCGGGUG-AGUGGAUCACG--UUGCCCAGGAUCUGUUCCAUUU--UUUAUUUUUUCUCCUCGUU-- ......(((((....((((((....))))))...))))).(-((..((((.(.--.......)))))..))).....--...................-- ( -26.20, z-score = -1.12, R) >droMoj3.scaffold_6680 18428048 92 + 24764193 GUACUUCCUCCAAAAGGCUAUACCCGUGGCCAUGGCGGGUG-AGUGGAUCAAA---UGUUUCUUUUUUUUUCUAUCU--UUAACUAUAUAUUCUCGCU-- ......((.(((...((((((....)))))).))).))(((-((((((..(((---.........)))..))))...--.............))))).-- ( -16.59, z-score = -0.10, R) >consensus GUACUUCCUGCAGAAGGCUAUUCCCGUGGCCAUGGCAGGUG_AGUUUGCUAAG__UUAAGCAGGGUGAUUUAUAUUU__UAAACUUUUUCAUUCCUUU__ ...((.(((((....((((((....))))))...)))))...))........................................................ (-12.72 = -12.63 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:33 2011