| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,875,173 – 9,875,264 |
| Length | 91 |
| Max. P | 0.985027 |

| Location | 9,875,173 – 9,875,264 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 59.93 |
| Shannon entropy | 0.76576 |
| G+C content | 0.49897 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -11.48 |
| Energy contribution | -10.20 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985027 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
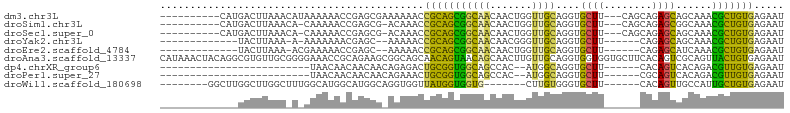
>dm3.chr3L 9875173 91 - 24543557 ----------CAUGACUUAAACAUAAAAAACCGAGCGAAAAAACCGCAGCGGCAACAACUGGUUGCAGGUGCUU---CAGCAGAGCAGCAAACGCUGUGAGAAU ----------..................................((((((((((((.....))))).(.(((((---.....))))).)...)))))))..... ( -25.00, z-score = -1.59, R) >droSim1.chr3L 9258803 89 - 22553184 ----------CAUGACUUAAACA-CAAAAACCGAGCG-ACAAACCGCAGCGGCAACAACUGGUUGCAGGUGCUU---CAGCAGAGCGGCAAACGCUGUGAGAAU ----------.............-.............-......((((((((((((.....))))).(.(((((---.....))))).)...)))))))..... ( -24.30, z-score = -0.68, R) >droSec1.super_0 2107512 89 - 21120651 ----------CAUGACUUAAACA-CAAAAACCGAGCG-ACAAACCGCAGCGGCAACAACUGGUUGCAGGUGCUU---CAGCAGAGCAGCAAACGCUGUGAGAAU ----------.............-.............-......((((((((((((.....))))).(.(((((---.....))))).)...)))))))..... ( -25.00, z-score = -1.18, R) >droYak2.chr3L 9851425 81 - 24197627 -------------UACUUAAA-A-AAAAAAACGAGC--AAAAACCGCAGCGGCAACAACGGGUUGCAGGUGCUU------CAGAGCAGCAAACGCUGUGAGAAU -------------........-.-............--......((((((((((((.....))))).(.((((.------...)))).)...)))))))..... ( -24.00, z-score = -2.49, R) >droEre2.scaffold_4784 9865427 82 - 25762168 -------------UACUUAAA-ACGAAAAACCGAGC--AAAAACCGCAGCGGCAACAACUGGUUGCAGGUGCUU------CAGAGCAUCAAACGCUGUGAGAAU -------------........-.((......))...--......((((((((((((.....))))).((((((.------...))))))...)))))))..... ( -26.00, z-score = -3.16, R) >droAna3.scaffold_13337 14805305 104 - 23293914 CAUAAACUACAGGCGUGUUGCGGGGAAACCGCAGAAGCGGCAGCAACAGUAACAGCAACUUGUUGCAGGUGGUGGUGCUUCACAGUCGCAGUUACUGUGAGAAU ((((((((...(((.((((((((.....)))))((((((.((.((...(((((((....)))))))...)).)).))))))))))))..))))..))))..... ( -36.10, z-score = -1.54, R) >dp4.chrXR_group6 5543779 71 + 13314419 -------------------------UAACAACAACAACAGAGACUGCGGUGGCAGCCAC--AUGGCAGGUGCUU------CACAGUCACAGACGUUGUGAGAAU -------------------------.....(((((..(.(.(((((.((.(((((((..--..)))...)))))------).))))).).)..)))))...... ( -20.00, z-score = -1.17, R) >droPer1.super_27 685049 71 - 1201981 -------------------------UAACAACAACAACAGAAACUGCGGUGGCAGCCAC--AUGGCAGGUGCUU------CGCAGUCACAGACGUUGUGAGAAU -------------------------.....(((((..(.(..(((((((.(((((((..--..)))...)))))------))))))..).)..)))))...... ( -21.70, z-score = -1.61, R) >droWil1.scaffold_180698 4980802 83 + 11422946 --------GGCUUGGCUUGGCUUUGGCAUGGCAUGGCAGGUGGUUAUGGUGGUG-------CUUGUGGGUGCUU------CACAGUUGCCAUUGCUGUGAGAAU --------((((......))))....(((((((((((((.((......(.((..-------((....))..)).------).)).)))))).)))))))..... ( -26.50, z-score = 0.34, R) >consensus _____________GACUUAAA_A_GAAAAACCGAGCG_AGAAACCGCAGCGGCAACAACUGGUUGCAGGUGCUU______CAGAGCAGCAAACGCUGUGAGAAU ............................................(((((((((((.......))))....((((........))))......)))))))..... (-11.48 = -10.20 + -1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:32 2011