| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,866,536 – 9,866,633 |
| Length | 97 |
| Max. P | 0.984936 |

| Location | 9,866,536 – 9,866,633 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 71.61 |
| Shannon entropy | 0.52309 |
| G+C content | 0.48282 |
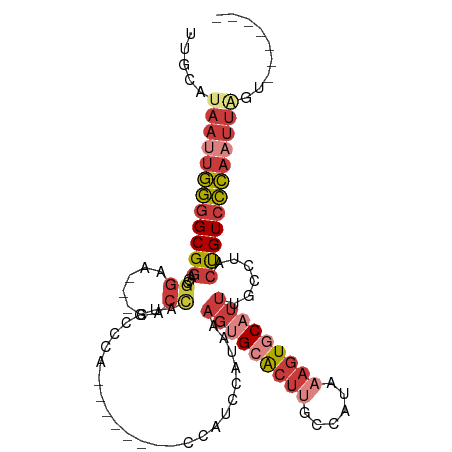
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -18.16 |
| Energy contribution | -20.24 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
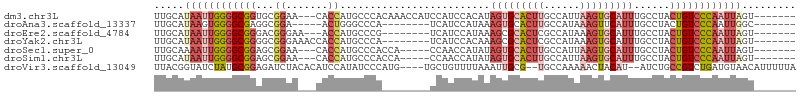
>dm3.chr3L 9866536 97 - 24543557 UUGCAUAAUUGGGGCGGUGCGGAA---CACCAUGCCCACAAACCAUCCAUCCACAUAGUGCACUUGCCAUUAAGUGCAUUUGCCUACUGUCCCAAUUAGU------- .....((((((((((((((((((.---..................)))........((((((((((....)))))))))).))..)))))))))))))..------- ( -31.41, z-score = -2.25, R) >droAna3.scaffold_13337 14796689 87 - 23293914 UUGCAUAAGUGGGGCGAGGCGGA-----ACUGGGCCCA--------UCAUCCAUAAAGUGCACUUGCCAUAAAGUUCAUUUGCCUACUGUCCCAAUUGGC------- ..((.....((((((.(((((((-----..((....))--------...)))...(((((.((((......)))).)))))))))...))))))....))------- ( -27.30, z-score = -0.68, R) >droEre2.scaffold_4784 9856392 89 - 25762168 UUGCAUAAUUGGGGCGGGACGGGAA---ACCAUGCCCG--------UCAUCCAUAAAGCGCACUCGCCAUAAAGUGCAUUUGCCUACUGUCCCAAUUAGU------- .....((((((((((((((((((..---......))))--------))...........(((((........))))).........))))))))))))..------- ( -35.60, z-score = -3.65, R) >droYak2.chr3L 9842595 92 - 24197627 UUGCAUAAUUGGGGCGGGGCGGGAAACCACCAUGCCCA--------UCAUCCACAAAGCGCACUCGCCAUAAAGUGCAUUUGCCUACUGUCCCAAUUAGU------- .....((((((((((((((.((....)).)).......--------.......((((..(((((........))))).))))....))))))))))))..------- ( -34.50, z-score = -2.64, R) >droSec1.super_0 2098984 92 - 21120651 UUGCAAAAUUGGGGCGGAGCGGAA---CACCAUGCCCACCA-----CCAACCAUAUAGUGCACUUGCCAUUAAGUGCAUUUGCCUACUGUCCCAAUUAGU------- ......(((((((((((.((((..---..))..........-----..........((((((((((....)))))))))).))...)))))))))))...------- ( -28.40, z-score = -1.75, R) >droSim1.chr3L 9250311 92 - 22553184 UUGCAUAAUUGGGGCGGAGCGGAA---CACCAUGCCCACCA-----CCAACCAUAUAGUGCACUUGCCAUUAAGUGCAUUUGCCUACUGUCCCAAUUAGU------- .....((((((((((((.((((..---..))..........-----..........((((((((((....)))))))))).))...))))))))))))..------- ( -29.80, z-score = -2.16, R) >droVir3.scaffold_13049 8996270 99 - 25233164 UUACGGUAUCUAUGCGGAGAUCUACACAUCCAUAUCCCAUG----UGCUGUUUUAAAUUGCG--UGCCAAAAACUACAU--AUCUGCCGUCUGAUGUAACAUUUUUA ..((((((..((((.(((..........)))........((----.(((((........)))--.)))).......)))--)..))))))................. ( -15.10, z-score = 0.54, R) >consensus UUGCAUAAUUGGGGCGGAGCGGAA___CACCAUGCCCA________CCAUCCAUAAAGUGCACUUGCCAUAAAGUGCAUUUGCCUACUGUCCCAAUUAGU_______ .....((((((((((((...((.......)).........................(((((((((......)))))))))......))))))))))))......... (-18.16 = -20.24 + 2.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:30 2011