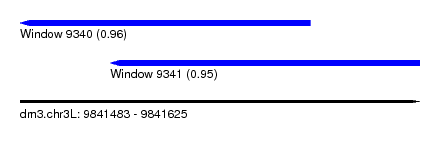
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,841,483 – 9,841,625 |
| Length | 142 |
| Max. P | 0.964610 |

| Location | 9,841,483 – 9,841,586 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 53.60 |
| Shannon entropy | 0.83874 |
| G+C content | 0.40053 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
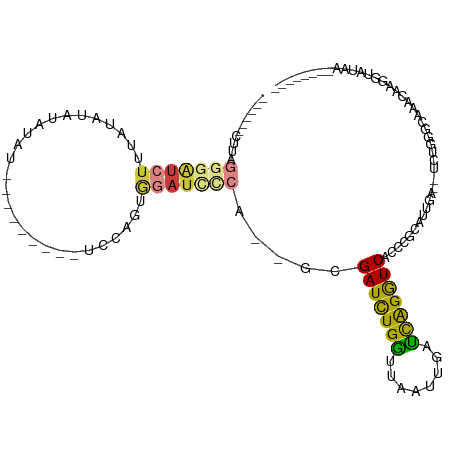
>dm3.chr3L 9841483 103 - 24543557 ------CCUAGGGAUCUUUAUAUACAUAUAUAUAUAUAUCCAGUGGAUUCCA--GCGAUCUGCUUAAUUGGGCAGGUCACCCUUAUUGGCCUCUGGGUAAACAAUGUAUAA-------- ------....................(((((((...(((((((.((......--..(((((((((....)))))))))..((.....)))).)))))))....))))))).-------- ( -28.50, z-score = -1.19, R) >droPer1.super_67 182405 100 + 321716 ----------------UUCGUUGAUAUAUGUAUGCAGAUAUCAUAGAUCAUGCAUAGAUCUGGUUAAUUG-CUGGAUCUCCUCUCCCAA--CCCGACCAUAUAUGCAUUAGCUGGUAAU ----------------...((((((.(((((((((((((.......))).)))..(((((..((.....)-)..)))))..........--.......))))))).))))))....... ( -22.50, z-score = -0.36, R) >dp4.chrXR_group5 594341 116 - 740492 UUCGUUGAUAUAUGUAUGUAUAUACAUAUGUAUGCAGAUAUCAUAGAUCAUGCAUAGAUCUGGUUUAUUG-CUGGAUCUCCUCUCCCAA--CCCGACCAUAUAUGCAUUAGCUGGUAAU ...((((((((((((((....))))))))))((((((((.......))).)))))(((((..((.....)-)..))))).......)))--)...((((.....((....))))))... ( -28.80, z-score = -0.84, R) >droEre2.scaffold_4784 9830747 92 - 25762168 ------CUUAGGGAUCUUUAUAUAU-----------------GUGGAUCCCA--GCGAUUUGGUUAAUUGAUCAGGUCACUCGCUUUGC--GCCGGGUAUACAAUAUAUAAAAAAAAGU ------..........(((((((((-----------------(((...(((.--(((((((((((....)))))))))....((...))--)).)))..))).)))))))))....... ( -24.50, z-score = -2.11, R) >droYak2.chr3L 9816445 89 - 24197627 ------CUUAGGGAUCGUUAUAUAU-----------------GUGGAUCCCA--GAGAUCUGGUUAAUUGAUCAGGUCACCCGCAUUUA--GCUGGGUAAACAAUGUAUAACAAAU--- ------....((((((.........-----------------...)))))).--..(((((((((....)))))))))((((((.....--)).))))..................--- ( -27.00, z-score = -2.73, R) >droSec1.super_0 2073922 93 - 21120651 ------CUUAGGGAUCUUUAUAUAUAUAU---------CCCAGUGGAUCUCA-GGCGAUCUGGGUAAUUGAUCAGGUCACCCGCAUUGG--UCUGGGCUAACAUGGUAUAA-------- ------....(((((.(........).))---------))).(((...((((-((((((.(((((.(((.....))).))))).))).)--))))))....))).......-------- ( -23.60, z-score = 0.25, R) >droSim1.chr3L 9224818 78 - 22553184 ------CUUAGGGAUCUUUAUAUAUAUAU---------CCCAGUGGAUCCCA--GCGAUCUGGUUAAUUGAUCAGGUCACCCGCAUUGG--UCUGGA---------------------- ------....(((((.(........).))---------)))........(((--(.(((((((((....)))))))))(((......))--))))).---------------------- ( -21.70, z-score = -0.48, R) >consensus ______CUUAGGGAUCUUUAUAUAUAUAU_________UCCAGUGGAUCCCA__GCGAUCUGGUUAAUUGAUCAGGUCACCCGCAUUGA__UCUGGGCAAACAAGGUAUAA________ ..........(((((((...........................))))))).....(((((((........)))))))......................................... (-11.22 = -11.23 + 0.01)

| Location | 9,841,515 – 9,841,625 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.43 |
| Shannon entropy | 0.66069 |
| G+C content | 0.39527 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -12.57 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
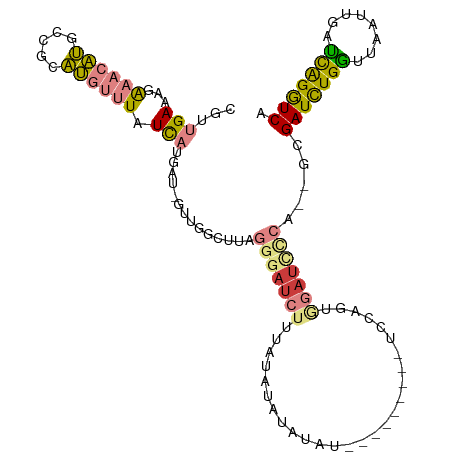
>dm3.chr3L 9841515 110 - 24543557 CGUUGAAAGAAACAUGCCGCAUGUUUAUCAUGAU-GUUUGCCUAGGGAUCUUUAUAUACAUAUAUAUAUAUAUCCAGUGGAUUCCA--GCGAUCUGCUUAAUUGGGCAGGUCA .((((....((((((....((((.....))))))-)))).((...((((...((((((....))))))...))))...))....))--))(((((((((....))))))))). ( -30.40, z-score = -1.61, R) >droPer1.super_67 182443 93 + 321716 UGCUGAAAUGUGUUGCCCGCCGCGUUGUU-UGUUCGUUGAU----------------AUAUGU--AUGCAGAUAUCAUAGAUCAUGCAUAGAUCUGGUUAAUUG-CUGGAUCU ((((((((((((........))))))...-...((..((((----------------((.((.--...)).))))))..))))).))).(((((..((.....)-)..))))) ( -23.40, z-score = -0.45, R) >dp4.chrXR_group5 594379 109 - 740492 UGCUGAAAUGUGUUGCCCGCCGCGUUGUU-UGUUCGUUGAUAUAUGUAUGUAUAUACAUAUGU--AUGCAGAUAUCAUAGAUCAUGCAUAGAUCUGGUUUAUUG-CUGGAUCU ((((((..((((..((.....))..((((-(((......((((((((((....))))))))))--..))))))).))))..))).))).(((((..((.....)-)..))))) ( -30.00, z-score = -1.00, R) >droEre2.scaffold_4784 9830785 93 - 25762168 CGUUGAAAGAAACAUGCCGCAUGUUUAUCAUGCU-GUUGGCUUAGGGAUCUUUAUAUAU-----------------GUGGAUCCCA--GCGAUUUGGUUAAUUGAUCAGGUCA .(((......)))..((((((((.....))))).-...)))...(((((((........-----------------..))))))).--..(((((((((....))))))))). ( -28.90, z-score = -2.39, R) >droYak2.chr3L 9816480 93 - 24197627 CGUUGAAAGAAACAUGCCGCAUGUUUAUCAUGAU-GUUUGCUUAGGGAUCGUUAUAUAU-----------------GUGGAUCCCA--GAGAUCUGGUUAAUUGAUCAGGUCA ......(((((((((....((((.....))))))-)))).))).((((((.........-----------------...)))))).--..(((((((((....))))))))). ( -26.20, z-score = -1.70, R) >droSec1.super_0 2073952 103 - 21120651 CGUUGAAAGAAACAUGCGGCAUGUUUAUCAAAAUGGUUGGCUUAGGGAUCUUUAUAUAUAUAU---------CCCAGUGGAUCUCA-GGCGAUCUGGGUAAUUGAUCAGGUCA ((((.....(((((((...))))))).....))))....((((.(((((((............---------......))))))))-)))(((((((........))))))). ( -26.57, z-score = -1.02, R) >droSim1.chr3L 9224834 101 - 22553184 CGUUGAAAGAAACAUGCCGCAUGUUUAUCAAAAU-GUUUGCUUAGGGAUCUUUAUAUAUAUAU---------CCCAGUGGAUCCCA--GCGAUCUGGUUAAUUGAUCAGGUCA ..((((...(((((((...))))))).))))...-....(((..(((((((............---------......))))))))--))(((((((((....))))))))). ( -29.27, z-score = -2.63, R) >consensus CGUUGAAAGAAACAUGCCGCAUGUUUAUCAUGAU_GUUGGCUUAGGGAUCUUUAUAUAUAUAU_________UCCAGUGGAUCCCA__GCGAUCUGGUUAAUUGAUCAGGUCA ...(((...((((((.....)))))).)))..............(((((((...........................))))))).....(((((((........))))))). (-12.57 = -12.83 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:28 2011