
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,710,395 – 4,710,491 |
| Length | 96 |
| Max. P | 0.535291 |

| Location | 4,710,395 – 4,710,491 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.69 |
| Shannon entropy | 0.51935 |
| G+C content | 0.45321 |
| Mean single sequence MFE | -17.03 |
| Consensus MFE | -6.19 |
| Energy contribution | -6.11 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4710395 96 + 23011544 -----------------GCACCCCCCACCAUCACCCCUUGCCAACAGUGAAAGCCCGGCUUU-----GUCUCUUUAUAUCUGCCGAGAUUUAAGCGUCAUAUAAAAUUUAUGCGUGUG -----------------((((.........((((............)))).......((((.-----(((((............)))))..))))(.((((.......))))))))). ( -14.40, z-score = -0.37, R) >droSim1.chr2L 4578872 93 + 22036055 --------------------CCCCCCACCAUCACCCUCUGCCGACGGUGAAAGCCCUGCUUU-----GUCUCUUUAUAUCUGCCGAGAUUUAAGCGUCAUAUAAAAUUUAUGCGUGUG --------------------.....((((((((((.((....)).))))).......((((.-----(((((............)))))..))))...............)).))).. ( -18.70, z-score = -2.27, R) >droSec1.super_5 2786837 94 + 5866729 -------------------CCCCCCCACCAUCACCCCCUGCCGACGGUGAAAGCCCUGCUUU-----GUCUCUUUAUAUCUGCCGAGAUUUAAGCGUCAUAUAAAAUUUAUGCGUGUG -------------------......((((((...........((((((....)))..((((.-----(((((............)))))..)))))))...........))).))).. ( -18.35, z-score = -2.36, R) >droYak2.chr2L 4717861 86 + 22324452 ---------------------------CCCUUUACUUUUGCCGGCAGUGAAAGCCCUGCUUU-----GUCUCUUUAUGUCUGCCGAGAUUUAAGCGUCAUAUAAAAUUUAUGCGUGUG ---------------------------..............((((((..((((..(......-----)...))))....))))))........(((.((((.......)))))))... ( -15.70, z-score = -0.55, R) >droEre2.scaffold_4929 4784959 93 + 26641161 --------------------AAAAAAACACGCCCCCUUUUCCGGCAGUGAAAGCCCUGCGUU-----GUCUCUUUAUGUCUGCCGAGAUUUAAGCGUCAUAUAAAAUUUAUGCGUGUG --------------------......((((((.........((((((..((((..(......-----)...))))....)))))).(((......))).............)))))). ( -20.40, z-score = -1.93, R) >droAna3.scaffold_12943 1910175 95 - 5039921 ------------------ACGUACGUCCCCGUAUCACCCCCAAUCCGCAAAAAGUCUGUUUU-----GUGUCUUUAUCUCUGCCGAGAUUUAAGCGUCAUAUAAAAUUUAUGCGUGUG ------------------..(((((....)))))...........((((....((..(((((-----(((((((.(((((....)))))..)))....)))))))))..))...)))) ( -15.70, z-score = -1.94, R) >droWil1.scaffold_180703 1447392 118 - 3946847 CCCCACUGCUCCUCCUCCUCCUCCCAUCACUCUUCACUCCCGUAUAUCUUUUUCUCUUUUUUAUUUGGUGUCUUUAUCUCUGCCGAGAUUUUAGCAUCAUAUAAAAUUUAUGUGCUUG .........................................((((((...........((((((.((((((....(((((....)))))....)))))).))))))...))))))... ( -15.94, z-score = -3.91, R) >consensus ____________________CCCCCCACCCUCACCCCUUGCCGACAGUGAAAGCCCUGCUUU_____GUCUCUUUAUAUCUGCCGAGAUUUAAGCGUCAUAUAAAAUUUAUGCGUGUG ...................................................................(((((............)))))....(((.((((.......)))))))... ( -6.19 = -6.11 + -0.08)

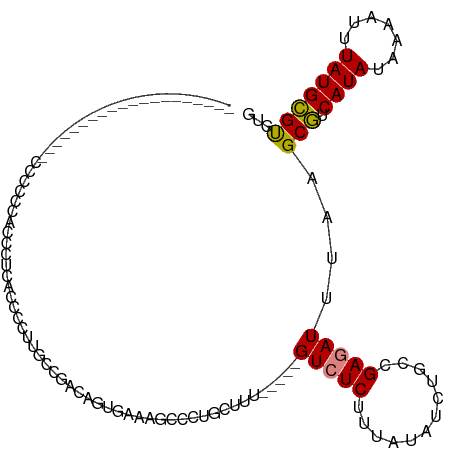
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:14 2011