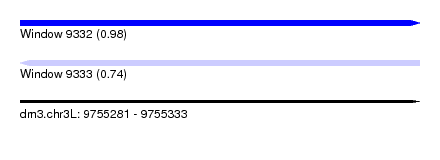
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,755,281 – 9,755,333 |
| Length | 52 |
| Max. P | 0.983590 |

| Location | 9,755,281 – 9,755,333 |
|---|---|
| Length | 52 |
| Sequences | 9 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 65.11 |
| Shannon entropy | 0.65138 |
| G+C content | 0.37430 |
| Mean single sequence MFE | -8.26 |
| Consensus MFE | -6.19 |
| Energy contribution | -6.13 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
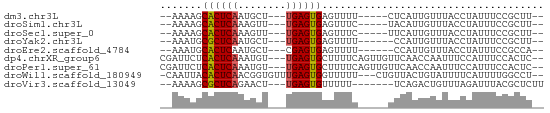
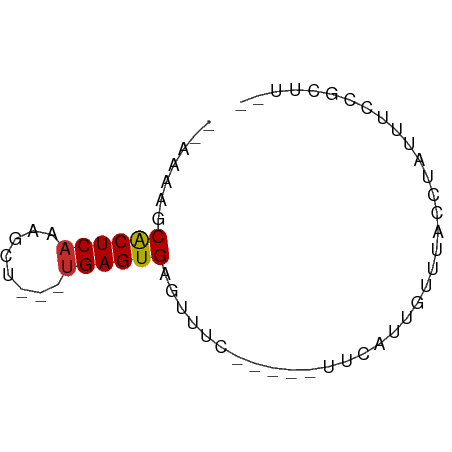
>dm3.chr3L 9755281 52 + 24543557 --AAAAGCACUCAAUGCU---UGAGUGAGUUUU-----CUCAUUGUUUACCUAUUUCCGCUU-- --..(((((((((.....---)))))(((....-----))).................))))-- ( -9.70, z-score = -1.78, R) >droSim1.chr3L 9137423 52 + 22553184 --AAAAGCACUCAAAGUU---UGAGUGAGUUUC-----UACAUUGUUUACCUAUUUCCGCUU-- --(((..((((((.....---))))))..))).-----........................-- ( -7.50, z-score = -0.85, R) >droSec1.super_0 1988915 52 + 21120651 --AAAAGCACUCAAAGUU---UGAGUGAGUUUC-----UUCAUUGUUUACCUAUUUCCGCUU-- --(((..((((((.....---))))))..))).-----........................-- ( -7.50, z-score = -0.92, R) >droYak2.chr3L 9727678 51 + 24197627 --AAAUGCGCUCAAUGCU---UGAGUGAGUUUU------CCAUUGUUUACCUAUUUCCGCUU-- --((((.((((((.....---)))))).)))).------.......................-- ( -6.90, z-score = -0.53, R) >droEre2.scaffold_4784 9743061 51 + 25762168 --AAAUGCACUCAAUGCU---CGAGUGAGUUUU------CCAUUGUUUACCUAUUUCCGCCA-- --((((.(((((......---.))))).)))).------.......................-- ( -6.10, z-score = -0.56, R) >dp4.chrXR_group6 3409050 59 - 13314419 CGAUUCUCACUCAAAUGU---UGAGUGCUUUUCAGUUGUUCAACCAAUUUCCAUUUCCACUC-- .((....((((((.....---))))))....))(((((......))))).............-- ( -8.20, z-score = -1.35, R) >droPer1.super_61 164845 59 - 350175 CGAUUCUCACUCAAAUGU---UGAGUGCUUUUCAGUUGUUCAACCAAUUUCCAUUUCCACUC-- .((....((((((.....---))))))....))(((((......))))).............-- ( -8.20, z-score = -1.35, R) >droWil1.scaffold_180949 5211681 58 - 6375548 -CAAUUACACUCAACGGUGUUUGAGUGGUUUUU---CUGUUACUGUAUUUUCAUUUUGGCCU-- -......(((((((......)))))))......---..........................-- ( -8.40, z-score = -0.14, R) >droVir3.scaffold_13049 16526293 52 - 25233164 --AAAAGCGCUCAGAACU---UGAGUGUUUUU-------UCAGACUGUUUAGAUUUACGCUCUU --((((((((((((...)---)))))))))))-------..(((.(((........))).))). ( -11.80, z-score = -1.06, R) >consensus __AAAAGCACUCAAAGCU___UGAGUGAGUUUC_____UUCAUUGUUUACCUAUUUCCGCUU__ .......((((((........))))))..................................... ( -6.19 = -6.13 + -0.06)

| Location | 9,755,281 – 9,755,333 |
|---|---|
| Length | 52 |
| Sequences | 9 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 65.11 |
| Shannon entropy | 0.65138 |
| G+C content | 0.37430 |
| Mean single sequence MFE | -7.76 |
| Consensus MFE | -4.94 |
| Energy contribution | -5.07 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.29 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
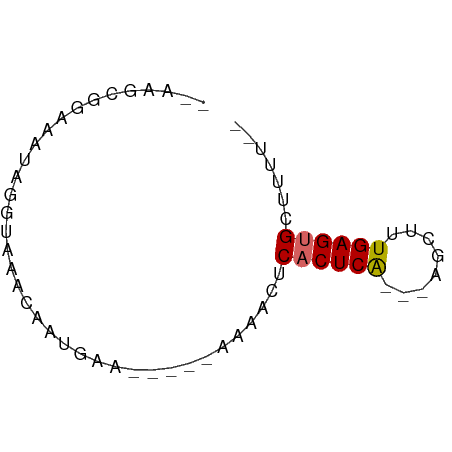
>dm3.chr3L 9755281 52 - 24543557 --AAGCGGAAAUAGGUAAACAAUGAG-----AAAACUCACUCA---AGCAUUGAGUGCUUUU-- --((((.................(((-----....)))(((((---.....)))))))))..-- ( -8.80, z-score = -0.79, R) >droSim1.chr3L 9137423 52 - 22553184 --AAGCGGAAAUAGGUAAACAAUGUA-----GAAACUCACUCA---AACUUUGAGUGCUUUU-- --........................-----......((((((---.....)))))).....-- ( -6.10, z-score = 0.35, R) >droSec1.super_0 1988915 52 - 21120651 --AAGCGGAAAUAGGUAAACAAUGAA-----GAAACUCACUCA---AACUUUGAGUGCUUUU-- --((((.................((.-----.....))(((((---.....)))))))))..-- ( -6.30, z-score = 0.13, R) >droYak2.chr3L 9727678 51 - 24197627 --AAGCGGAAAUAGGUAAACAAUGG------AAAACUCACUCA---AGCAUUGAGCGCAUUU-- --..(((.................(------(....)).((((---.....)))))))....-- ( -5.80, z-score = 0.21, R) >droEre2.scaffold_4784 9743061 51 - 25762168 --UGGCGGAAAUAGGUAAACAAUGG------AAAACUCACUCG---AGCAUUGAGUGCAUUU-- --............(((..(((((.------....(((....)---)))))))..)))....-- ( -6.90, z-score = 0.05, R) >dp4.chrXR_group6 3409050 59 + 13314419 --GAGUGGAAAUGGAAAUUGGUUGAACAACUGAAAAGCACUCA---ACAUUUGAGUGAGAAUCG --...........((..(..(((....)))..)....((((((---.....))))))....)). ( -9.80, z-score = -0.60, R) >droPer1.super_61 164845 59 + 350175 --GAGUGGAAAUGGAAAUUGGUUGAACAACUGAAAAGCACUCA---ACAUUUGAGUGAGAAUCG --...........((..(..(((....)))..)....((((((---.....))))))....)). ( -9.80, z-score = -0.60, R) >droWil1.scaffold_180949 5211681 58 + 6375548 --AGGCCAAAAUGAAAAUACAGUAACAG---AAAAACCACUCAAACACCGUUGAGUGUAAUUG- --..........................---......(((((((......)))))))......- ( -7.60, z-score = -0.82, R) >droVir3.scaffold_13049 16526293 52 + 25233164 AAGAGCGUAAAUCUAAACAGUCUGA-------AAAAACACUCA---AGUUCUGAGCGCUUUU-- ((((((((...((..(((....(((-------........)))---.)))..))))))))))-- ( -8.70, z-score = -0.56, R) >consensus __AAGCGGAAAUAGGUAAACAAUGAA_____AAAACUCACUCA___AGCUUUGAGUGCUUUU__ .....................................((((((........))))))....... ( -4.94 = -5.07 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:21 2011