| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,743,245 – 9,743,325 |
| Length | 80 |
| Max. P | 0.895894 |

| Location | 9,743,245 – 9,743,325 |
|---|---|
| Length | 80 |
| Sequences | 12 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 70.04 |
| Shannon entropy | 0.68912 |
| G+C content | 0.57227 |
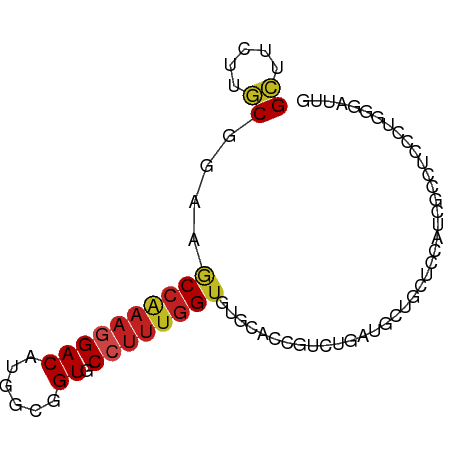
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -11.21 |
| Energy contribution | -11.41 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.895894 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
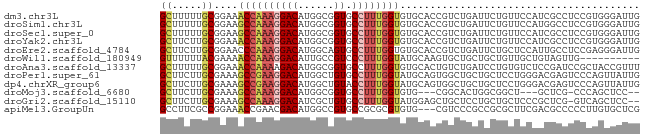
>dm3.chr3L 9743245 80 + 24543557 GCUUUUUGCGGAAACCAAAGGACAUGGCGGUGCCUUUGGUGUGCACCGUCUGAUUCUGUUCCAUCGCCUCCGUGGGAUUG ((.....))((((.....((((((.(((((((((......).)))))))))).)))).))))(((.((.....))))).. ( -26.70, z-score = -1.16, R) >droSim1.chr3L 9125046 80 + 22553184 GCUUUUUGCGGAAGCCAAAGGACAUGGCGGUGCCUUUGGUGUGCACCGUCUGAUUCUGUUCCAUGGCCUCCGUGGGAUUG ...(((..((((.((((..(((((.(((((((((......).))))))))......)))))..)))).))))..)))... ( -35.80, z-score = -3.47, R) >droSec1.super_0 1976903 80 + 21120651 GCUUUUUGCGGAAGCCAAAGGACAUGGCGGUGCCUUUGGUGUGCACCGUCUGAUUCUGUUCCAUCGCCUCCGUGGGAUUG ...(((..((((.((....(((((.(((((((((......).))))))))......)))))....)).))))..)))... ( -30.00, z-score = -1.83, R) >droYak2.chr3L 9715082 80 + 24197627 GCUUCUUGCGGAAACCAAAGGACAUGGCGGUGCCUUUGGUGUGCACCGUCUGAUUCUGUUCCAUCGCCUCCGUGGGAUUG ...(((..((((.......(((((.(((((((((......).))))))))......))))).......))))..)))... ( -29.14, z-score = -1.92, R) >droEre2.scaffold_4784 9730171 80 + 25762168 GCUUCUUGCGGAACCCAAAGGACAUGGCAGUGCCUUUGGUGUGCACCGUCUGAUUCUGCUCCAUUGCCUCCGAGGGAUUG ((.((..((((.(((((((((.(((....)))))))))).))...))))..))....))(((.(((....))).)))... ( -24.60, z-score = -0.38, R) >droWil1.scaffold_180949 5197729 70 - 6375548 GUUUUUUACGAAAACCAAAGGACAUUGCCGUCCCUUUGGUAUGCAAGUGCUGCUGCUGUUGCUGUAGUUG---------- .....(((((...((((((((((......)).))))))))..((((..((....))..)))))))))...---------- ( -19.70, z-score = -1.81, R) >droAna3.scaffold_13337 19657513 80 - 23293914 GCUUUUUGCGGAAACCAAAAGACAUGGCGGUGCCUUUGGUGUGCACUGUCUGAUCCUGUGUCUCCGAUCCGCUACCGUUU .......(((((.......(((((((((((((((......).)))))))).......))))))....)))))........ ( -23.61, z-score = -1.47, R) >droPer1.super_61 152471 80 - 350175 GCUUCUUGCGAAAGCCGAAGGACAUGGCUGUGCCUUUGGUAUGCAGUGGCUGCUGCUCCUGGGACGAGUCCCAGUUAUUG ((.....))....((((((((.(((....)))))))))))..(((((....)))))..((((((....))))))...... ( -31.40, z-score = -2.07, R) >dp4.chrXR_group6 3396850 80 - 13314419 GCUUCUUGCGAAAGCCGAAGGACAUGGCUGUACCUUUGGUAUGCAGUGGCUGCUGCUCCUGGGACGAGUCCCAGUUAUUG ((.....))....(((((((((((....))).))))))))..(((((....)))))..((((((....))))))...... ( -30.80, z-score = -2.26, R) >droMoj3.scaffold_6680 6626837 71 + 24764193 GCUUCUUGCGAAAGCCAAAGGACAUGGCGGUGCCUUUGGUGUG---CGGCACUGGCGGCU---GCUCG-CCCAGCUCC-- (((....((((..((((((((.(((....)))))))))))..(---((((.......)))---)))))-)..)))...-- ( -27.40, z-score = 0.14, R) >droGri2.scaffold_15110 20103890 77 - 24565398 GCUUCUUGCGAAAGCCAAAGGACAUCGCUGUGCCUUUGGUAUGGAGCUGCUCCUGCUGCUCCCGCUCG-GUCAGCUCC-- (((((..(((...(((((((((((....))).))))))))..(((((.((....)).))))))))..)-)..)))...-- ( -31.10, z-score = -2.56, R) >apiMel3.GroupUn 104458878 77 + 399230636 GCCUUCGCCGGAAACCGAACGACAUGGCCGUGCCGCGCGUGUG---CGUCCCGCCGCGCUUCGACGCCCCCUUGUGCUCG ((.((((..(....)))))..(((.((..(((.((.(((((.(---((...))))))))..)).)))..)).)))))... ( -22.40, z-score = 1.35, R) >consensus GCUUCUUGCGGAAGCCAAAGGACAUGGCGGUGCCUUUGGUGUGCACCGUCUGAUGCUGCUCCAUCGCCUCCCUGGGAUUG ((.....))....((((((((((......)).))))))))........................................ (-11.21 = -11.41 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:18 2011