| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,691,363 – 4,691,467 |
| Length | 104 |
| Max. P | 0.865240 |

| Location | 4,691,363 – 4,691,467 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.55354 |
| G+C content | 0.42678 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -10.67 |
| Energy contribution | -10.98 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.865240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
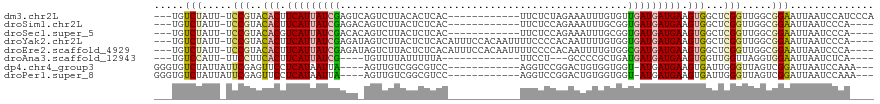
>dm3.chr2L 4691363 104 - 23011544 ---UGUCUAUU-UCCGUACACUUCAUUAUCGAGUCAGUCUUACACUCAC------------UUCUCUAGAAAUUUGUGUUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAUCCAUCCCA ---.(((....-.(((..(((((((((((((((.....)))((((....------------(((....)))....)))).))))))))))))...)))..)))(((.....)))...... ( -30.00, z-score = -2.87, R) >droSim1.chr2L 4557874 100 - 22036055 ---UGUCUAUU-UCCGUACACUUCAUUAUCGAGACAGUCUUACUCUCAC------------UUCUCCAGAAAUUUGCGGUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAUCCCA---- ---.(((....-.(((..((((((((((((((((.........))))..------------(((....))).........))))))))))))...)))..)))(((.....)))..---- ( -28.40, z-score = -1.91, R) >droSec1.super_5 2769037 100 - 5866729 ---UGUCUAUU-UCCGUACACGUCAUUAUCGACACAGUCUUACUCUCAC------------UUCUCCAGAAAUUUGCGGUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAUCCCA---- ---.(((....-.(((..(((.(((((((((((...)))..........------------(((....))).........)))))))).)))...)))..)))(((.....)))..---- ( -22.00, z-score = 0.16, R) >droYak2.chr2L 4698455 112 - 22324452 ---UGUCUAUU-UCCGUACACUUCAUUAUCGAGAUAGUCUUACUCUCACAUUUCCACAAUUUUCCCCACAAUUUUGUGGUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAUCCCA---- ---.(((....-.(((..((((((((((((((((.........))))......((((((..((......))..)))))).))))))))))))...)))..)))(((.....)))..---- ( -35.00, z-score = -4.05, R) >droEre2.scaffold_4929 4763374 112 - 26641161 ---UGUCUAUU-UCCGUACACUUCAUUAUCGAGAUAGUCUUACUCUCACAUUUCCACAAUUUUCCCCACAAUUUUGUGGCGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAUCCCA---- ---.(((....-.(((..((((((((((((((((.........))))......((((((..((......))..)))))).))))))))))))...)))..)))(((.....)))..---- ( -35.10, z-score = -4.23, R) >droAna3.scaffold_12943 1892549 92 + 5039921 ---UGUCCAUU-UUCCUUCACUUCAUUAUCG----UGUUUUAUUUUUA-------------UUCCU---GCCCCGCUGAUGAUGAUGAAGUGGUUGGUUAGGUGGAAUUAAUCUCA---- ---..((((((-(.((.((((((((((((((----(............-------------.....---.........)))))))))))))))..))..)))))))..........---- ( -26.27, z-score = -3.62, R) >dp4.chr4_group3 536755 100 + 11692001 GGGUGUCUAUUAUUCGAGUUCCUCAUAAUUA----AGUUGUCGGCGUCC------------AGGUCCGGACUGUGGUGGU-AUGAUGAAGUGAUUGGGUUAGUCGGAUUAAUCCAAA--- ((((.......((((((.(..((((..((((----..(..(((.(..((------------((.......))).)..)..-.)))..)..))))))))..).))))))..))))...--- ( -24.10, z-score = -0.04, R) >droPer1.super_8 1580179 100 + 3966273 GGGUGUCUAUUAUUCGAGUUCCUCAUAAUUA----AGUUGUCGGCGUCC------------AGGUCCGGACUGUGGUGGU-AUGAUGAAGUGAUUGGGUUAGUCGGAUUAAUCCAAA--- ((((.......((((((.(..((((..((((----..(..(((.(..((------------((.......))).)..)..-.)))..)..))))))))..).))))))..))))...--- ( -24.10, z-score = -0.04, R) >consensus ___UGUCUAUU_UCCGUACACUUCAUUAUCGAGA_AGUCUUACUCUCAC____________UUCUCCAGAAUUUUGUGGUGAUGAUGAAGUGGCUCGGUUGGCGGAAUUAAUCCCA____ .....(((.....(((..(((.(((((((((................................................))))))))).)))...))).....))).............. (-10.67 = -10.98 + 0.31)
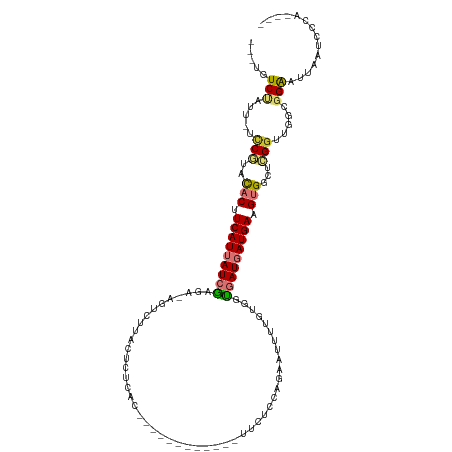
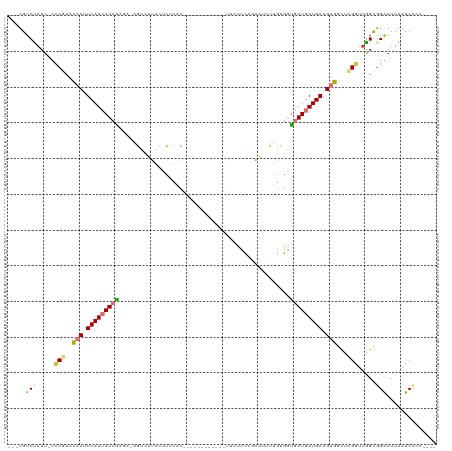
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:17:13 2011