
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,713,929 – 9,714,034 |
| Length | 105 |
| Max. P | 0.585005 |

| Location | 9,713,929 – 9,714,034 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.90 |
| Shannon entropy | 0.71405 |
| G+C content | 0.48311 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -9.23 |
| Energy contribution | -8.00 |
| Covariance contribution | -1.23 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
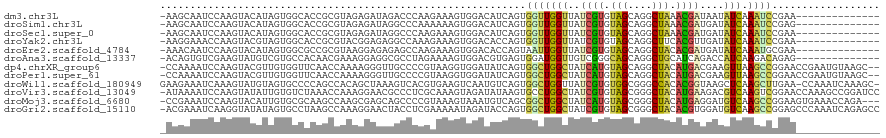
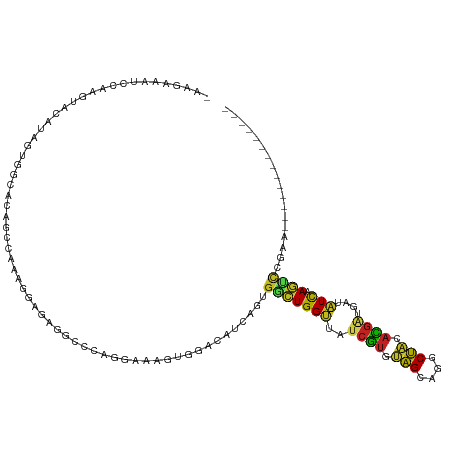
>dm3.chr3L 9713929 105 - 24543557 -AAGCAAUCCAAGUACAUAGUGGCACCGCGUAGAGAUAGACCCAAGAAAGUGGACAUCAGUGGUUGGUUAUCGUGUAGCAGGCUAAACGAUAAUAUCAAAUCCGAA-------------- -.......((((.(((...(((....))).....(((....(((......)))..))).))).))))(((((((.(((....))).))))))).............-------------- ( -22.10, z-score = -0.52, R) >droSim1.chr3L 9094521 105 - 22553184 -AAGCAAUCCAAGUACAUAGUGGCACCGCGUAGAGAUAGGCCCAAAAAAGUGGACAUCAGUGGUUGGUUAUCGUGUAGCAGGCUAAACGAUGAUAUCAAAUCCGAG-------------- -.......((((.(((...(((....))).....(((.(..(((......))).)))).))).))))(((((((.(((....))).))))))).............-------------- ( -23.60, z-score = -0.49, R) >droSec1.super_0 1947563 105 - 21120651 -AAGCAAUCCAAGUACAUAGUGGCACCGCGUAGAGAUAGGCCCAAGAAAGUGGACAUCAGUGGUUGGUUAUCGUGUAGCAGGCUAAACGAUGAUAUCAAAUCCGAA-------------- -.......((((.(((...(((....))).....(((.(..(((......))).)))).))).))))(((((((.(((....))).))))))).............-------------- ( -23.60, z-score = -0.26, R) >droYak2.chr3L 21978803 105 + 24197627 -AAGGAAACCAAGUACGUAGUGGCACCGCGUACGGAGAGGCCAAAGAAAGUGGACACCAGUGGUUGGUUAUCGUGUAGCAGGCUUCACGUUGAUAUCAAAUCCGAA-------------- -..(((((((((.(((....((((.(((....)))....)))).......(((...)))))).))))))..((((.((....)).))))...........)))...-------------- ( -28.10, z-score = -0.47, R) >droEre2.scaffold_4784 9701014 105 - 25762168 -AAACAAUCCAAGUACAUAGUGGCGCCGCGUAAGGAGAGAGCCAAGAAAGUGGACACCAGUAAUUGGUUAUCGUGUAGCAGGCUACACGAUGAUAUCAAAUGCGAA-------------- -......((((.........((((.((......)).....))))......)))).....(((.(((((((((((((((....))))))))))))..))).)))...-------------- ( -31.26, z-score = -3.02, R) >droAna3.scaffold_13337 19629107 105 + 23293914 -ACAGUGUCGAAGUAUGUCGUGCCACAACGAAAGGAGGCGCCUAGAAAAGUGGACGUGAGUGGAUGGUUGUCGGGCAGCAGGCUGCAUCAGACCAUCAAGACAGAG-------------- -....((((....(((((((((((....(....)..)))))((.....))..))))))....(((((((.....((((....))))....)))))))..))))...-------------- ( -34.90, z-score = -1.76, R) >dp4.chrXR_group6 3367282 117 + 13314419 -CCAAAAUCCAAGUACGUUGUGGUUCAACCAAAAGGGUUGCCCCGUAAGGUGGAUAUCAGUGGCUGGCUAUCAUGUAGCAGGCUACAUGACGAAGUUAAGCCGGAACCGAAUGUAAGC-- -............((((((.((((((((((.....)))).((((....)).)).......(((((((((.((((((((....))))))))...)))).)))))))))))))))))...-- ( -39.80, z-score = -2.84, R) >droPer1.super_61 123092 117 + 350175 -CCAAAAUCCAAGUACGUUGUGGUUCAACCAAAAGGGUUGCCCCGUAAGGUGGAUAUCAGUGGCUGGCUAUCAUGUAGCAGGCUACAUGACGAAGUUAAGCCGGAACCGAAUGUAAGC-- -............((((((.((((((((((.....)))).((((....)).)).......(((((((((.((((((((....))))))))...)))).)))))))))))))))))...-- ( -39.80, z-score = -2.84, R) >droWil1.scaffold_180949 5165933 118 + 6375548 GAAGAAAUCAAAGUAUGUAGUGCCCCAGCCACAGCUAAAGUCACGUGAAGUCAAUGUCAGUGGCUGGUUAUCGUGUGGCGGGCCACACGGUAAGCUCAAGCUUGAA-CCAAAUCAAAGC- .......((((.((.((.(((...((((((((.((....)).((((.......))))..))))))))(((((((((((....)))))))))))))))).)))))).-............- ( -38.60, z-score = -2.89, R) >droVir3.scaffold_13049 16483162 119 + 25233164 -AUAAAAUCCAAGUAUAUUGUGUCUAAACCAAAGGAACGCCCUCGCAAAGUAGAUAUAAGUGCCUGGCUAUCGUGUAGCGGGCUACAUGAAGACGUCAAGUCGGAACCAAAGCCGGAUCC -.....((((..((((.(((((((((......(((.....))).......)))))))))))))..((((.((((((((....)))))))).(((.....)))........)))))))).. ( -35.82, z-score = -3.01, R) >droMoj3.scaffold_6680 6595234 116 - 24764193 -CCGAAAUCCAAGUACAUUGUGCGCAAGCCAAGCGAGCAGCCCCGUAAAGUAAAUGUCAGCGGCUGGCUAUCAUGUAGCGGGCUACAUGAGGAUGUCAAGCCGGAAGUGAAACCAGA--- -......(((.......(((.((....)))))((..(((....(.....)....)))..))(((((((..((((((((....))))))))....))).)))))))............--- ( -34.70, z-score = -0.86, R) >droGri2.scaffold_15110 20069675 119 + 24565398 -ACGAAAUCAAGGUAUAUAGUGCCUAAGCCAAAGGAACUACCUCGAAAAAUAGAUACCAGUGGCUGGCUAUCGUGUAGCGGGCUACACGUGGAUGUCAAGCCGGAGCCCAAAUCAGAGCC -.((...((.((((....(((.(((.......))).))))))).))......(((((((((((((.(((((...))))).))))))...))).))))....))..((.(......).)). ( -29.90, z-score = 0.13, R) >consensus _AAGAAAUCCAAGUACAUAGUGGCACAGCCAAAGGAGAGGCCCAGGAAAGUGGACAUCAGUGGCUGGUUAUCGUGUAGCAGGCUACACGAUGAUAUCAAGUCCGAA______________ .............................................................(((((((..((((.(((....))).))))....))).)))).................. ( -9.23 = -8.00 + -1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:13 2011