
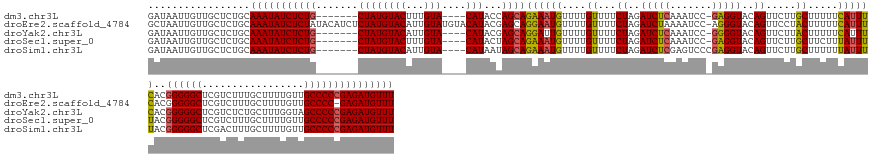
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,689,173 – 9,689,322 |
| Length | 149 |
| Max. P | 0.966642 |

| Location | 9,689,173 – 9,689,322 |
|---|---|
| Length | 149 |
| Sequences | 5 |
| Columns | 161 |
| Reading direction | forward |
| Mean pairwise identity | 90.12 |
| Shannon entropy | 0.16671 |
| G+C content | 0.39961 |
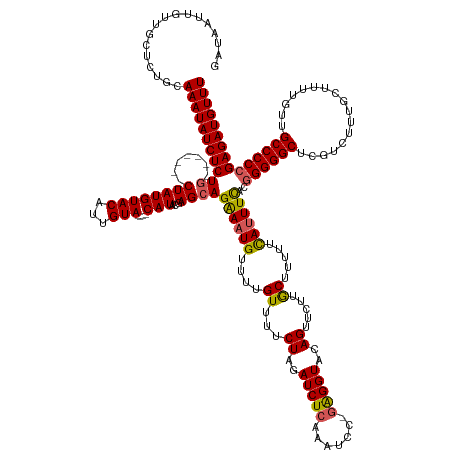
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -29.19 |
| Energy contribution | -28.59 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
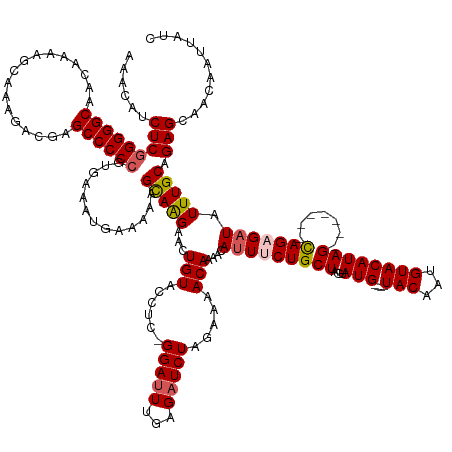
>dm3.chr3L 9689173 149 + 24543557 GAUAAUUGUUGCUCUGCAAAUAUCUCUG-------CUAUGUACUUUGUA----CAUACCAGCAGAAAUGUUUUGUUUUCUAGAUCUCAAAUCC-GAGGUACAGUUCUUGCUUUUUCAUUUCACGGGGGCUCGUCUUUGCUUUUGUUGCCCCCGAGAUGUUU .................(((((((((((-------((((((((...)))----)))...))))((((((....((...((..(((((......-)))))..)).....)).....))))))..((((((.................))))))))))))))) ( -40.63, z-score = -2.91, R) >droEre2.scaffold_4784 9672959 159 + 25762168 GCUAAUUGUUGCUCUGCAAAUAUCUCUAUACAUCUCUAUGUACAUUGUAUGUACAUACGAGCAGGAAUGUUUUGUUUUCUAGAUCUAAAAUCC-AGGGUACAGUUCCUACUUUUUCAUUUCACGGGGGCUCGUCUUUGCUUUUGUUGCCCC-GAGAUGUUU ((........)).....(((((((((.......((((((((((((...))))))))).))).((((((....(((.((((.(((.....))).-)))).)))))))))................(((((.................)))))-))))))))) ( -39.63, z-score = -2.07, R) >droYak2.chr3L 21953460 149 - 24197627 GAUAAUUGUUGCUCUGCAAAUAUCUCUG-------CUAUGUACAUUGUA----CAUACGAGCAGGAUUGUUUUGUUUUCUAGAUCUCAAAUCC-GGGGUACAGUUCUUACUUUUUCAUUUCACGGGGGCUCGUCUCUGCUUUGGUAGCCCCCGAGAUGUUU ...((((((.((((((((((.(((.(((-------((((((((...)))----)))...))))))))...)))).......(((.....))))-)))))))))))..........((((((..(((((((.(((........))))))))))))))))... ( -40.60, z-score = -1.78, R) >droSec1.super_0 1922742 149 + 21120651 GAUAAUUGUUGCUCUGCAAAUAUCUCUG-------CUAUGUACUUUGUA----CAUACUAGCAGAAAUGUUUUGUUUUCUAGAUCUCAAAUCC-GAGGUACAGUUCUUGCUUCUUUAUUUUACGGGGGCUCGUCUUUGCUUUUGUUGCCCCCGAGAUGUUU ((((.((((......)))).))))((((-------((((((((...)))----))...))))))).................(((((......-((((((.......))))))..........((((((.................))))))))))).... ( -40.83, z-score = -3.27, R) >droSim1.chr3L 9066888 150 + 22553184 GAUAAUUGUUGCUCUGCAAAUAUCUCUG-------CUAUGUACAUUGUA----CAUAAUAGCAGAAAUGUUUUGUUUUCUAGAUCUCGAGUCCCGAGGUACAGUUCUUGCUUUUUUAUUUUACGGGGGCUCGACUUUGCUUUUGUUGCCCCCGAGAUGUUU ((((.((((......)))).))))((((-------((((((((...)))----))...)))))))(((((((.((...((..((((((.....))))))..)).....))............(((((((.(((........)))..)))))))))))))). ( -41.40, z-score = -2.74, R) >consensus GAUAAUUGUUGCUCUGCAAAUAUCUCUG_______CUAUGUACAUUGUA____CAUACGAGCAGAAAUGUUUUGUUUUCUAGAUCUCAAAUCC_GAGGUACAGUUCUUGCUUUUUCAUUUCACGGGGGCUCGUCUUUGCUUUUGUUGCCCCCGAGAUGUUU .................(((((((((.....................................((((((....((...((..(((((.......)))))..)).....)).....))))))..((((((.................))))))))))))))) (-29.19 = -28.59 + -0.60)
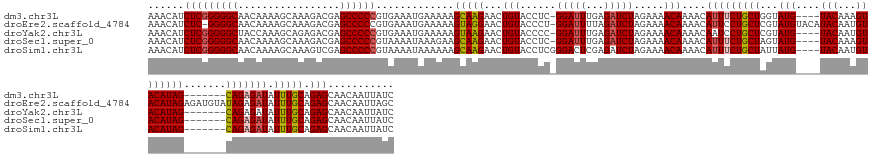
| Location | 9,689,173 – 9,689,322 |
|---|---|
| Length | 149 |
| Sequences | 5 |
| Columns | 161 |
| Reading direction | reverse |
| Mean pairwise identity | 90.12 |
| Shannon entropy | 0.16671 |
| G+C content | 0.39961 |
| Mean single sequence MFE | -39.11 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9689173 149 - 24543557 AAACAUCUCGGGGGCAACAAAAGCAAAGACGAGCCCCCGUGAAAUGAAAAAGCAAGAACUGUACCUC-GGAUUUGAGAUCUAGAAAACAAAACAUUUCUGCUGGUAUG----UACAAAGUACAUAG-------CAGAGAUAUUUGCAGAGCAACAAUUAUC .....((.(((((((.................))))))).)).........(((((..(((...(((-(....))))...)))..........((((((((((((((.----......)))).)))-------))))))).)))))............... ( -38.63, z-score = -3.10, R) >droEre2.scaffold_4784 9672959 159 - 25762168 AAACAUCUC-GGGGCAACAAAAGCAAAGACGAGCCCCCGUGAAAUGAAAAAGUAGGAACUGUACCCU-GGAUUUUAGAUCUAGAAAACAAAACAUUCCUGCUCGUAUGUACAUACAAUGUACAUAGAGAUGUAUAGAGAUAUUUGCAGAGCAACAAUUAGC ..(((((((-(((((.................))))).............((((((((.(((...((-(((((...)))))))...))).....))))))))..(((((((((...))))))))))))))))..........((((...))))........ ( -41.33, z-score = -3.66, R) >droYak2.chr3L 21953460 149 + 24197627 AAACAUCUCGGGGGCUACCAAAGCAGAGACGAGCCCCCGUGAAAUGAAAAAGUAAGAACUGUACCCC-GGAUUUGAGAUCUAGAAAACAAAACAAUCCUGCUCGUAUG----UACAAUGUACAUAG-------CAGAGAUAUUUGCAGAGCAACAAUUAUC .....((.((((((((..(........)...)))))))).)).................(((.....-(((((....................)))))(((((.((((----(((...)))))))(-------((((....))))).))))))))...... ( -38.55, z-score = -3.12, R) >droSec1.super_0 1922742 149 - 21120651 AAACAUCUCGGGGGCAACAAAAGCAAAGACGAGCCCCCGUAAAAUAAAGAAGCAAGAACUGUACCUC-GGAUUUGAGAUCUAGAAAACAAAACAUUUCUGCUAGUAUG----UACAAAGUACAUAG-------CAGAGAUAUUUGCAGAGCAACAAUUAUC ......(((((((((.................)))))).............(((((..(((...(((-(....))))...)))..........((((((((((((((.----......)))).)))-------))))))).))))).)))........... ( -37.53, z-score = -3.65, R) >droSim1.chr3L 9066888 150 - 22553184 AAACAUCUCGGGGGCAACAAAAGCAAAGUCGAGCCCCCGUAAAAUAAAAAAGCAAGAACUGUACCUCGGGACUCGAGAUCUAGAAAACAAAACAUUUCUGCUAUUAUG----UACAAUGUACAUAG-------CAGAGAUAUUUGCAGAGCAACAAUUAUC ......(((((((((.................)))))).............(((((..(((...((((.....))))...)))..........((((((((((...((----(((...))))))))-------))))))).))))).)))........... ( -39.53, z-score = -3.92, R) >consensus AAACAUCUCGGGGGCAACAAAAGCAAAGACGAGCCCCCGUGAAAUGAAAAAGCAAGAACUGUACCUC_GGAUUUGAGAUCUAGAAAACAAAACAUUUCUGCUAGUAUG____UACAAUGUACAUAG_______CAGAGAUAUUUGCAGAGCAACAAUUAUC ....(((((((((((.................))))))(((..(((....((((.(((.(((......(((((...)))))..........))).)))))))..))).....)))....................)))))..((((...))))........ (-23.26 = -23.34 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:12 2011