
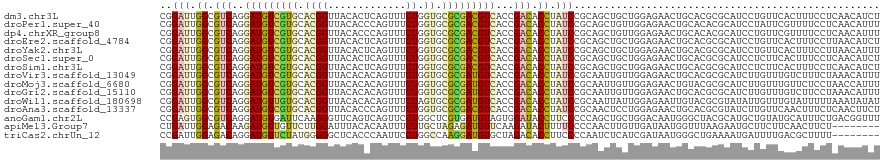
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,661,968 – 9,662,128 |
| Length | 160 |
| Max. P | 0.814574 |

| Location | 9,661,968 – 9,662,088 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Shannon entropy | 0.50319 |
| G+C content | 0.51591 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.58 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
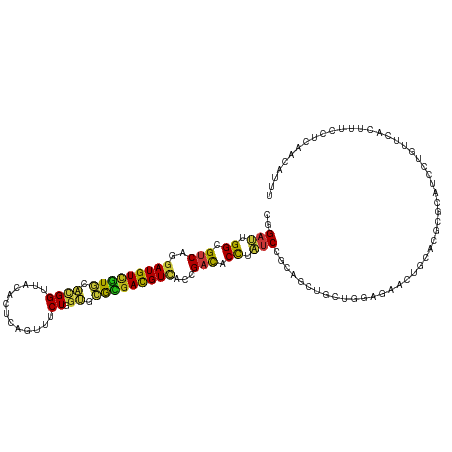
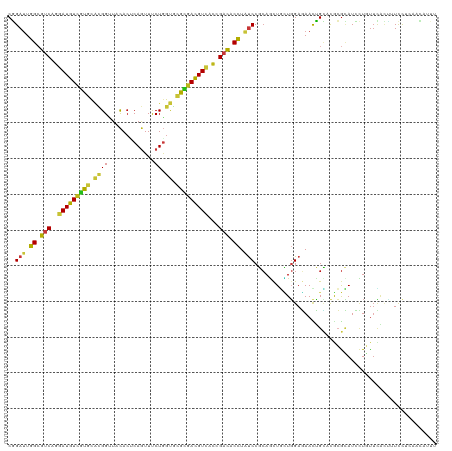
>dm3.chr3L 9661968 120 - 24543557 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUGGAGAACUGCACGCGCAUCCUGUUCACUUUCCUCAACAUCU .((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))((.(((((.(.....).))).))))......................... ( -44.00, z-score = -2.19, R) >droPer1.super_40 41921 120 + 810552 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGUUGGAGAACUGCACACGCAUCCUAUUCGUUUUCCUCAACAUUU (((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).)))))....((((((((((((((....)))........)))))))..))))... ( -46.60, z-score = -3.68, R) >dp4.chrXR_group8 653064 120 + 9212921 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGUUGGAGAACUGCACACGCAUCCUGUUCGUUUUCCUCAACAUUU (((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).)))))....((((((((((((((....)))........)))))))..))))... ( -46.60, z-score = -3.14, R) >droEre2.scaffold_4784 9645720 120 - 25762168 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUGGAGAACUGCACGCGCAUCCUGUUCACUUUCCUUAACAUCU .((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))((.(((((.(.....).))).))))......................... ( -44.00, z-score = -2.36, R) >droYak2.chr3L 21926418 120 + 24197627 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUGGAGAACUGCACGCGCAUCCUGUUCACUUUCCUUAACAUUU .((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))((.(((((.(.....).))).))))......................... ( -44.00, z-score = -2.52, R) >droSec1.super_0 1895712 120 - 21120651 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUGGAGAACUGCACGCGCAUCCUCUUCACUUUCCUCAACAUCU .((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))((....))((((((..(((....)))...))))))............... ( -44.70, z-score = -2.88, R) >droSim1.chr3L 9039365 120 - 22553184 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUGGAGAACUGCACGCGCAUCCUCUUCACUUUCCUCAACAUCU .((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))((....))((((((..(((....)))...))))))............... ( -44.70, z-score = -2.88, R) >droVir3.scaffold_13049 14928787 120 + 25233164 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUGGAGAACUGCACGCGCAUCUUGUUUGUCUUUCUAAACAUUU (((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))....((((..((((..(((.(((.....))).)))..)))).))))... ( -42.90, z-score = -2.61, R) >droMoj3.scaffold_6680 10481256 120 - 24764193 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUGGAGAACUGUACGCGCAUCUUGUUUGUUCUCCUAACCAUUU (((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))).....((((((((((...(((.......)))..))))))).)))..... ( -46.50, z-score = -3.56, R) >droGri2.scaffold_15110 8941529 120 - 24565398 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUGGAGAACUGCACGCGCAUCUUGUUUGUCUUCCUAAACAUUU (((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))....(((((((((...(((.........)))...)))))...))))... ( -40.60, z-score = -1.79, R) >droWil1.scaffold_180698 1532723 120 + 11422946 CGGAUUGGCGUCAGGAUGUUGUGCACGGUUACACACAGUUUCUAGUGCGCGAUGUCACCGACACCUAUCCGCAAUUAUUGGAGAAUUGUACGCGUAUAUUGUUUGUAUUUUUAAAUAUAU .((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))((((((.......))))))....(((((((.............))))))) ( -36.02, z-score = -2.05, R) >droAna3.scaffold_13337 3915181 120 - 23293914 CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAACUCCUGGAGAACUGCACGCGUAUCUUGUUCAACUUUCUCAACUUCU (((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).))))).........(((((......(((.....)))......)))))....... ( -42.40, z-score = -2.79, R) >anoGam1.chr2L 44307890 120 + 48795086 CCGAGUGGCGUCAGGAUGUGAUUCAAGGGUUCAGUCAGUUCCUGGCUCGUGAUGUAGUGGAUACCUUCCCCCAGCUGCUGGACAAUGGGCUACGCAUGCUGUAUGCAUUUCUGACGGUUU (((.((((((((((((..(((((.........)))))..)))))))..((...((((((((.....)))....)))))...)).....)))))(((((...)))))........)))... ( -35.40, z-score = 1.09, R) >apiMel3.Group7 6496384 112 + 9974240 CUGAUUGGAGACAAGAUGUUGUUCUUGGAUUUACACAAUUUCUUGCUAGAGAUGUUCAAGAUACUUUUCCCCAACUUGUUGAUAAUGGUUUAAGAAUGCUUCUUCAACUUCU-------- ...((((..((((((.((..((((((((((.......((((((....)))))))))))))).)).......)).))))))..))))((((.((((.....)))).))))...-------- ( -20.01, z-score = 0.59, R) >triCas2.chrUn_12 480922 112 + 931581 CCGAUUGGAGACAGGAUGUUCUAUGGGGGCUCACCCAAUUCCUGGCCAAGGAUGUGCUAGACACCUUCCCCAAUCUCAUCGAUAAUGGGCUGAAAAUGAUUUUGACGCUUUU-------- ..((((((.((.(((.(((....((((......))))....(((((((....)).))))))))))))).))))))...........((((...(((....)))...))))..-------- ( -28.10, z-score = 0.77, R) >consensus CGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUGGAGAACUGCACGCGCAUCCUGUUCACUUUCCUCAACAUUU ..(((.((.(((..(((((((((.((((.............)).)).)))))))))...))).)).)))................................................... (-16.80 = -16.58 + -0.22)
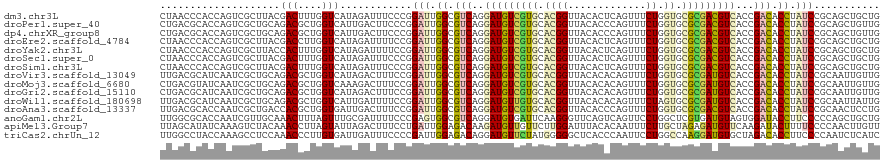
| Location | 9,662,008 – 9,662,128 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.44500 |
| G+C content | 0.53611 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -18.12 |
| Energy contribution | -17.91 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9662008 120 - 24543557 CUAACCCACCAGUCGCUUACGACUUUGGUCAUAGAUUUCCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUG .........((((.(((...(((....)))..........(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))).))).)))) ( -47.00, z-score = -3.70, R) >droPer1.super_40 41961 120 + 810552 CUGACGCACCAGUCGCUGCAGACGCUGGUCAUUGACUUCCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGUUG ..(((......)))(((((.(((....)))...........((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).)))))))))..... ( -52.30, z-score = -3.10, R) >dp4.chrXR_group8 653104 120 + 9212921 CUGACGCACCAGUCGCUGCAGACGCUGGUCAUUGACUUCCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGUUG ..(((......)))(((((.(((....)))...........((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).)))))))))..... ( -52.30, z-score = -3.10, R) >droEre2.scaffold_4784 9645760 120 - 25762168 CUAACCCACCAGUCGCUUACGACCUUGGUCAUAGAUUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUG .........((((.(((...(((....)))..........(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))).))).)))) ( -48.10, z-score = -4.02, R) >droYak2.chr3L 21926458 120 + 24197627 CUAACCCACCAGUCGCUUACCACUUUGGUCAUAGAUUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUG .........((((.(((.(((.....)))...........(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))).))).)))) ( -45.70, z-score = -3.74, R) >droSec1.super_0 1895752 120 - 21120651 CUAACCCACCAGUCGCUUACGACUUUGGUCAUAGAUUUCCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUG .........((((.(((...(((....)))..........(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))).))).)))) ( -47.00, z-score = -3.70, R) >droSim1.chr3L 9039405 120 - 22553184 CUAACCCACCAGUCGCUUACGACUUUGGUCAUAGAUUUCCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUG .........((((.(((...(((....)))..........(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))).))).)))) ( -47.00, z-score = -3.70, R) >droVir3.scaffold_13049 14928827 120 + 25233164 UUGACGCAUCAAUCGCUGCAGACGCUGGUCAUAGACUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUG .((((((.......)).((....))..))))..(((.((.(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))).))..))). ( -41.20, z-score = -1.21, R) >droMoj3.scaffold_6680 10481296 120 - 24764193 CUGACGUAUCAAUCGCUGCAGACGCUGGUCAAAGACUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUG .........((((.((....(((....)))...........((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))).))))... ( -40.40, z-score = -1.09, R) >droGri2.scaffold_15110 8941569 120 - 24565398 CUGACGCAUCAAUCGCUGCAGACGCUGGUCAUAGACUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUG ..(((((.......))(((.(((....)))...........((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))))...))). ( -41.20, z-score = -1.11, R) >droWil1.scaffold_180698 1532763 120 + 11422946 UUGACGCAUCAAUCGCUGCAGACGCUGGUCAUUGAUUUUCCGGAUUGGCGUCAGGAUGUUGUGCACGGUUACACACAGUUUCUAGUGCGCGAUGUCACCGACACCUAUCCGCAAUUAUUG .....((((((((.(((((....)).))).)))))).....((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))))........ ( -41.90, z-score = -2.42, R) >droAna3.scaffold_13337 3915221 120 - 23293914 UUGACGCACCAAUCGCUGACCACGCUGGUGAUUGACUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAACUCCUG .....((..((((((((.........)))))))).......((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).))))))........ ( -47.80, z-score = -3.38, R) >anoGam1.chr2L 44307930 120 + 48795086 UUGGCGCACCAAUCGUUGCAAACUUUAGUUUGCGAUUUUCCCGAGUGGCGUCAGGAUGUGAUUCAAGGGUUCAGUCAGUUCCUGGCUCGUGAUGUAGUGGAUACCUUCCCCCAGCUGCUG ....(((.(((.(((((((((((....))))))))......))).))).(((((((..(((((.........)))))..)))))))..)))..((((((((.....)))....))))).. ( -35.40, z-score = -0.39, R) >apiMel3.Group7 6496416 120 + 9974240 UUAGCAUAUCAAAGUCUACAAACCUUAGUAUUAGACUUUCCUGAUUGGAGACAAGAUGUUGUUCUUGGAUUUACACAAUUUCUUGCUAGAGAUGUUCAAGAUACUUUUCCCCAACUUGUU ...(((((((((((((((...((....))..)))))))).....(((....))))))).)))((((((((.......((((((....))))))))))))))................... ( -22.11, z-score = -0.13, R) >triCas2.chrUn_12 480954 120 + 931581 UUGGCCUACCAAAGCCUCCAAACCCUUGUGAUUGAUUUCCCCGAUUGGAGACAGGAUGUUCUAUGGGGGCUCACCCAAUUCCUGGCCAAGGAUGUGCUAGACACCUUCCCCAAUCUCAUC ((((((......(((((((((((.(((((..(..(((.....)))..)..)))))..)))...))))))))............))))))(((.(((.....)))..)))........... ( -30.77, z-score = 0.67, R) >consensus CUGACGCACCAAUCGCUGCAGACGCUGGUCAUAGAUUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUG ....................(((....)))............(((.((.(((..(((((((((.((((.............)).)).)))))))))...))).)).)))........... (-18.12 = -17.91 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:09 2011