
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,628,506 – 9,628,609 |
| Length | 103 |
| Max. P | 0.835948 |

| Location | 9,628,506 – 9,628,609 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 66.91 |
| Shannon entropy | 0.65191 |
| G+C content | 0.33301 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -7.20 |
| Energy contribution | -7.07 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
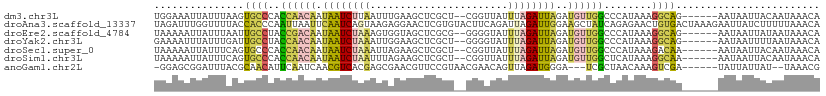
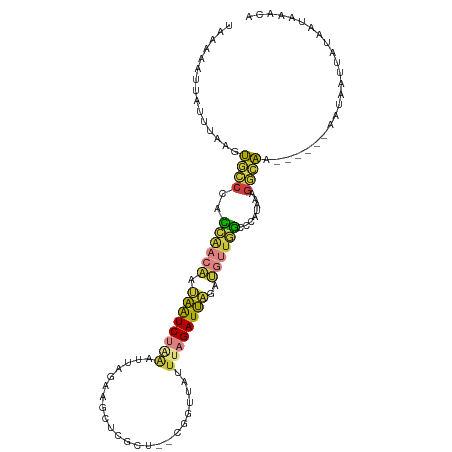
>dm3.chr3L 9628506 103 + 24543557 UGGAAAUUAUUUAAGUGCCCACCAACAAUAAUCUUAAUUUGAAGCUCGCU--CGGUUAUUUAGAUUAGAUGUUGGCCCAUAAAGGCAG------AAUAAUUACAAUAAACA ..(.((((((((...((((..((((((.((((((....((((.(....))--)))......))))))..))))))........)))))------))))))).)........ ( -19.00, z-score = -0.88, R) >droAna3.scaffold_13337 3872357 111 + 23293914 UAGAUUUGGUUUUACCACCCAAUUAAUUCAAUCAGUAAGAGGAACUCGUGUACUUCAGAUUAGAUUGGAAGCUAUCAGAGAACUGUGACUAAAGAAUUAUCUUUUUAAACA ..((((.(((......))).))))..((((((((((..((((.((....)).))))..))).)))))))...............((....(((((....)))))....)). ( -17.70, z-score = 0.56, R) >droEre2.scaffold_4784 9612609 103 + 25762168 UAAAAAUUAUUUAAUUGCCUACCGACAAUAAUCUAAAGUGGUAGCUCGCG--GGGGUAUUUAGAUUAGAUGUUGGCCCAUAAAGGCAG------AAUAAUUAUAAUAAACA ....((((((((...(((((.((((((.(((((((((......((((...--.)))).)))))))))..)))))).......))))))------))))))).......... ( -24.50, z-score = -1.97, R) >droYak2.chr3L 21892939 103 - 24197627 GAAAAUUUAUUUGAUUGCCUACCAACAAUAAUCUAAAUUGGAAGCUCGCU--GGGGUAUUUAGAUUAGAUGUUGGCCCAUAAAGGCAG------AAUAAUUUUAAUAAACA ..(((.((((((...(((((.((((((.((((((((((.....((((...--.))))))))))))))..)))))).......))))))------))))).)))........ ( -24.50, z-score = -2.18, R) >droSec1.super_0 1862417 103 + 21120651 UAAAAAUUAUUUCAGUGCCCACCAACAAUAAUCUAAAUUAGAAGCUCGCU--CGGUUAUUUAGAUUAGAUGUUGGCCCAUAAAGACAA------AAUAAUUACAAUAAACA ....((((((((.........((((((.((((((((((..((.(....))--)....))))))))))..))))))............)------))))))).......... ( -17.80, z-score = -2.36, R) >droSim1.chr3L 9003401 103 + 22553184 UAAAAAUUAUUUCAGUGCCCACCAACAAUAAUCUAAUUUAGAAGCUCGCU--CGGUUAUUUAGAUUAGAUGUUGGCUCAUAAAGGCAA------AAUAAUUACAAUAAACA ....((((((((...((((..((((((....(((((((((((((((....--.)))).)))))))))))))))))........)))))------))))))).......... ( -22.40, z-score = -3.34, R) >anoGam1.chr2L 44285081 99 - 48795086 -GGAGCGGAUUUACGCAACAUUCAAUCAACGUCACGAGCGAACGUUCCGUAACGAACAGUUAGAUGGGA---UCGCUAACAAAGUCGA------UAUUAUUAU--UAAACG -(..(((......)))..).....(((.((......(((((...(((((((((.....)))..))))))---)))))......)).))------)........--...... ( -19.60, z-score = -0.33, R) >consensus UAAAAAUUAUUUAAGUGCCCACCAACAAUAAUCUAAAUUAGAAGCUCGCU__CGGUUAUUUAGAUUAGAUGUUGGCCCAUAAAGGCAA______AAUAAUUAUAAUAAACA ...............((((..((((((.((((((((.......................))))))))..))))))........))))........................ ( -7.20 = -7.07 + -0.13)
| Location | 9,628,506 – 9,628,609 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 66.91 |
| Shannon entropy | 0.65191 |
| G+C content | 0.33301 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -9.12 |
| Energy contribution | -8.76 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
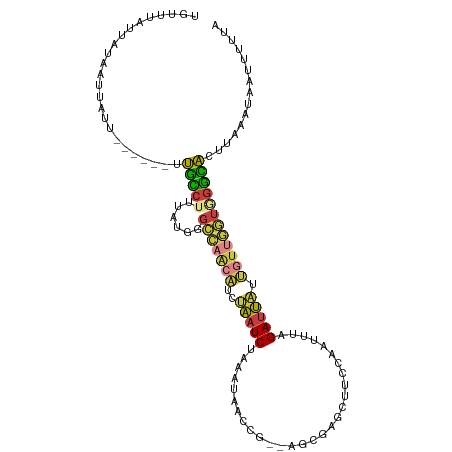
>dm3.chr3L 9628506 103 - 24543557 UGUUUAUUGUAAUUAUU------CUGCCUUUAUGGGCCAACAUCUAAUCUAAAUAACCG--AGCGAGCUUCAAAUUAAGAUUAUUGUUGGUGGGCACUUAAAUAAUUUCCA ..........(((((((------.(((((......(((((((..((((((.(((....(--((....)))...))).)))))).))))))))))))....))))))).... ( -20.90, z-score = -1.07, R) >droAna3.scaffold_13337 3872357 111 - 23293914 UGUUUAAAAAGAUAAUUCUUUAGUCACAGUUCUCUGAUAGCUUCCAAUCUAAUCUGAAGUACACGAGUUCCUCUUACUGAUUGAAUUAAUUGGGUGGUAAAACCAAAUCUA .........(((((((((.(((((.(.((..(((((...(((((...........))))).)).)))...)).).)))))..))))))....(((......)))...))). ( -16.50, z-score = 0.61, R) >droEre2.scaffold_4784 9612609 103 - 25762168 UGUUUAUUAUAAUUAUU------CUGCCUUUAUGGGCCAACAUCUAAUCUAAAUACCCC--CGCGAGCUACCACUUUAGAUUAUUGUCGGUAGGCAAUUAAAUAAUUUUUA ..........(((((((------.(((((......(((.(((..(((((((((......--(....).......))))))))).))).))))))))....))))))).... ( -18.62, z-score = -1.01, R) >droYak2.chr3L 21892939 103 + 24197627 UGUUUAUUAAAAUUAUU------CUGCCUUUAUGGGCCAACAUCUAAUCUAAAUACCCC--AGCGAGCUUCCAAUUUAGAUUAUUGUUGGUAGGCAAUCAAAUAAAUUUUC .................------.(((((......(((((((..((((((((((.....--((....))....)))))))))).))))))))))))............... ( -22.40, z-score = -2.35, R) >droSec1.super_0 1862417 103 - 21120651 UGUUUAUUGUAAUUAUU------UUGUCUUUAUGGGCCAACAUCUAAUCUAAAUAACCG--AGCGAGCUUCUAAUUUAGAUUAUUGUUGGUGGGCACUGAAAUAAUUUUUA ..........(((((((------((((((......(((((((..((((((((((....(--((....)))...)))))))))).))))))))))))...)))))))).... ( -23.50, z-score = -2.27, R) >droSim1.chr3L 9003401 103 - 22553184 UGUUUAUUGUAAUUAUU------UUGCCUUUAUGAGCCAACAUCUAAUCUAAAUAACCG--AGCGAGCUUCUAAAUUAGAUUAUUGUUGGUGGGCACUGAAAUAAUUUUUA ..........(((((((------((((((......(((((((..((((((((......(--((....))).....)))))))).))))))))))))...)))))))).... ( -23.80, z-score = -2.53, R) >anoGam1.chr2L 44285081 99 + 48795086 CGUUUA--AUAAUAAUA------UCGACUUUGUUAGCGA---UCCCAUCUAACUGUUCGUUACGGAACGUUCGCUCGUGACGUUGAUUGAAUGUUGCGUAAAUCCGCUCC- ......--.((((((..------......))))))(((.---...((......))....(((((.((((((((.(((......))).)))))))).)))))...)))...- ( -20.20, z-score = -0.15, R) >consensus UGUUUAUUAUAAUUAUU______UUGCCUUUAUGGGCCAACAUCUAAUCUAAAUAACCG__AGCGAGCUUCCAAUUUAGAUUAUUGUUGGUGGGCACUUAAAUAAUUUUUA ........................(((((......(((((((..(((((.............................))))).))))))))))))............... ( -9.12 = -8.76 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:04 2011