
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,619,003 – 9,619,117 |
| Length | 114 |
| Max. P | 0.554431 |

| Location | 9,619,003 – 9,619,117 |
|---|---|
| Length | 114 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.73 |
| Shannon entropy | 0.64966 |
| G+C content | 0.54990 |
| Mean single sequence MFE | -43.42 |
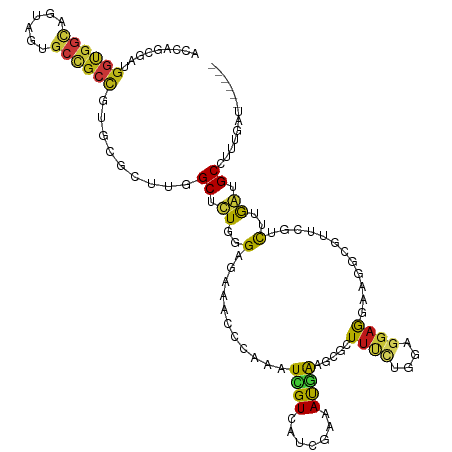
| Consensus MFE | -12.21 |
| Energy contribution | -11.47 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
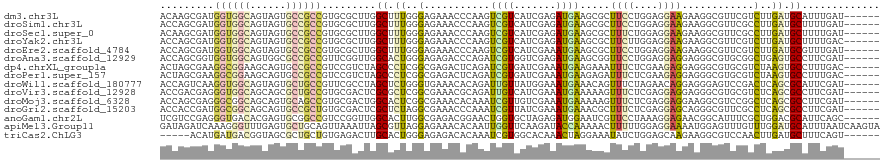
>dm3.chr3L 9619003 114 + 24543557 ACAAGCGAUGGUGGCAGUAGUGCCGCCGUGCGCUUGGCUUUGGGAGAAACCCAAGUCGUCAUCGAGAUGAAGCGCUUCCUGGAGGAAGAAGGCGUUCGUCUUGAUGCAUUUGAU------ .(((((((((((((((....))))))))).))))))(.((((((.....)))))).)(.((((((((((((((.(((((....)))))...)).))))))))))))).......------ ( -56.40, z-score = -5.53, R) >droSim1.chr3L 8994009 114 + 22553184 ACCAGCGAUGGUGGCAGUAGUGCCGCCGUGCGCUUGGCUUUGGGAGAAACCCAAGUCGUCAUCGAGAUGAAGCGCUUCCUGGAGGAAGAAGGCGUUCGCCUUGAUGCUUUUGAU------ ...(((((((((((((....))))))))).)))).(((((.(((.....))))))))(.(((((((.((((((.(((((....)))))...)).)))).)))))))).......------ ( -48.90, z-score = -2.94, R) >droSec1.super_0 1853087 114 + 21120651 ACAAGCGAUGGUGGCAGUAGUGCCGCCGUGCGCUUGGCUUUGGGAGAAACCCAAGUCGUCAUCGAGAUGAAGCGCUUUCUGGAGGAAGAAGGCGUUCGCCUUGAUGCUUUUGAU------ .(((((((((((((((....))))))))).))))))(.((((((.....)))))).)(.(((((((.((((.((((((((......)))))))))))).)))))))).......------ ( -54.40, z-score = -4.72, R) >droYak2.chr3L 21883520 114 - 24197627 ACCAGCGAUGGUGGCAGUAGUGCCGCCGUGCGCUUGGCUUUGGGAGAAACCCAAGUCGUCAUCGAGAUGAAGCGCUUCUUGGAGGAAGAAGGCGUUCGUCUUGAUGCUUUUGAC------ .(((((((((((((((....))))))))).))).)))..(((((.....)))))((((.((((((((((((((.((((((.....)))))))).))))))))))))....))))------ ( -54.50, z-score = -4.74, R) >droEre2.scaffold_4784 9603063 114 + 25762168 ACCAGCGAUGGUGGCAGUAGUGCCGCCGUGCGCUUGGCUUUGGGAGAAACCCAAGUCGUCAUCGAAAUGAAGCGCUUCCUGGAGGAAGAAGGCGUUCGUCUUGAUGCGUUUGAU------ .(((((((((((((((....))))))))).))).))).((((((.....)))))).((.((((((.(((((((.(((((....)))))...)).))))).))))))))......------ ( -48.60, z-score = -3.13, R) >droAna3.scaffold_12929 1765369 114 + 3277472 ACCAGCGGUGGUGGCAGUGGCGCCGCCGUUCGGUUGGCACUGGGAGAGACCCAGAUCGUGGUCGAGAUGAAGCGGUUCCUGGAGGAGGAGGGCGUGCGGCUGAGUGCCUUCGAU------ (((((((((((((.(...).)))))))))).))).((((((.((.(.(.(((..(((((..((.....)).)))))((((.....))))))))...)..)).))))))......------ ( -46.30, z-score = -0.17, R) >dp4.chrXL_group1a 8541500 114 - 9151740 ACUAGCGAAGGCGGAAGCAGUGCCGCCGUCCGUCUAGCCCUCGGCGAGACUCAGAUCGUGAUCGAAAUGAAGAAAUUUCUCGAAGAGGAGGGCGUGCGUCUAAGUGCCUUUGAC------ .....((((((((....)(((((((((.(((((((.(((...))).))))(((..(((....)))..)))................))).)))).))).))....)))))))..------ ( -41.00, z-score = -1.08, R) >droPer1.super_157 41370 114 - 72662 ACUAGCGAAGGCGGAAGCAGUGCCGCCGUCCGUCUAGCCCUCGGCGAGACUCAGAUCGUGAUCGAAAUGAAGAGAUUUCUCGAAGAGGAGGGCGUGCGUCUAAGUGCCUUUGAC------ .....((((((((....)(((((((((.(((((((.(((...))).))))(((..(((....)))..))).(((....))).....))).)))).))).))....)))))))..------ ( -43.50, z-score = -1.47, R) >droWil1.scaffold_180777 3014656 114 + 4753960 ACCAGUCAAGGUGGCAGUAGUGCUGCCGUUCGCCUAGCUCUGGGUGAAACACAGAUUGUUAUGGAAAUGAAACAGUUUCUAGAACAGGAGGGAGUCCGACUCAGCGCAUUCGAU------ .((((((..((..(((....)))..)).((((((((....)))))))).....))))(((.(((((((......))))))).)))....))((((.((......)).))))...------ ( -34.50, z-score = -0.66, R) >droVir3.scaffold_12928 7266967 114 - 7717345 ACCGACGAGGGUGGCAGCAGCGCUGCCGUGCGACUCGCGCUCGGCGAAACGCAGAUUGUCAUCGAAAUGAAAAAGUUUCUCGAGGAGGAGGGCGUGCGUCUCAGCGCCUUCGAU------ .((..(((.(((((((((.(((.(((((.(((.....))).)))))...))).).))))))))(((((......)))))))).))..((((((((........))))))))...------ ( -46.10, z-score = -0.49, R) >droMoj3.scaffold_6328 3044957 114 - 4453435 ACCAGCGAGGGCGGCAGCAGUGCAGCCGUGCGACUGGCACUCGGCGAAACACAAAUCGUUGUCGAAAUGAAAAAGUUUCUCGAGGAGGAAGGCGUCCGGCUCAGCGCCUUCGAU------ .....(((((((((((....)))(((((..((.((..(.(((((.(((((.....(((((.....)))))....))))))))))...)..))))..)))))...))))))))..------ ( -44.90, z-score = -1.40, R) >droGri2.scaffold_15203 5118954 114 - 11997470 ACCACCGAUGGCGGCAGCAGUGCCGCUGUGCGACUCGCUCUAGGCGAAACCCAAAUCGUUAUCGAAAUGAAACGCUUUCUCGAGGAGCAGGGCGUUCGCCUCAGCGCCUUCGAU------ .....(((.(((((((....))))((((.((((..((((((.(((((........))))).............(((((.....))))))))))).))))..))))))).)))..------ ( -43.40, z-score = -1.32, R) >anoGam1.chr2L 36237207 114 - 48795086 UCGUCCGAGGGUGACACGAGUGCGGCCGUCCGGUUGGCACUUGGCGAGACGGAACUGGUGCUAGAGAUGGAAUCGUUCCUAAAGGAGAACGGCAUUUCGCUGGACGCAUUCAGC------ (((((((..(....).(((((((((((....)))).))))))).)).)))))...........((((((...(((((((.....).))))))))))))((((((....))))))------ ( -39.50, z-score = -0.16, R) >apiMel3.Group11 12252151 120 - 12576330 GAUAGAUCAAAGGGUUUGAGUGCUGCAGUUAAAUUAGCGUUAGGAGAAACACAAUUGGUUCAAGAUACCAAAAACUUUUUGGAGGAAAAUGGAGUUUGUUUGGAUGCAUUUAAUCAAGUA ..((((((....))))))..((((...(((((((..(((((..((.((((.((.(((((.......)))))...(((....))).....))..)))).))..))))))))))))..)))) ( -22.10, z-score = -0.08, R) >triCas2.ChLG3 28351790 109 - 32080666 -----ACAUGAUGACGGUAGCGCUGCUGUGAGACUUGCACUGGGAGAGACACAAAUCGUGGCACAAACUAGGAAAUAUCUGGAGCAAGAAGGCGUCCAACUUGAUGCUUUCAGU------ -----.((((((....((..(.((.(((((.......))).)).)).)..))..)))))).......(((((.....))))).....(((((((((......)))))))))...------ ( -27.20, z-score = 0.34, R) >consensus ACCAGCGAUGGUGGCAGUAGUGCCGCCGUGCGCUUGGCUCUGGGAGAAACCCAAAUCGUCAUCGAAAUGAAGCGCUUUCUGGAGGAGGAAGGCGUUCGUCUUGAUGCCUUUGAU______ .........((((((......)))))).........((.((..(...........((((.......)))).....((((....))))............)..)).))............. (-12.21 = -11.47 + -0.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:02 2011