| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,598,213 – 9,598,303 |
| Length | 90 |
| Max. P | 0.855836 |

| Location | 9,598,213 – 9,598,303 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Shannon entropy | 0.49105 |
| G+C content | 0.53116 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -15.91 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9598213 90 + 24543557 AACUGGGCGCGUUCGUUUUCACGCCGCUUGUCAGGUCGUUGAAAAUGUUACGACCGCCUAUCCUUCCUUCC----UUGCAAACCUCACCGAACA- ...(((((((((.((((((((((((........)).)).))))))))..)))..))))))...........----...................- ( -22.50, z-score = -1.28, R) >droSim1.chr3L 8972956 90 + 22553184 AACUGGGCGCGUUCGUUUUCACGCCGCUUGUCAGGUCGUUGAAAAUGUUACGACCGCCUAUCCUUCCUUCC----UUGCAAACCUCCCCGAACA- ...(((((((((.((((((((((((........)).)).))))))))..)))..))))))...........----...................- ( -22.50, z-score = -1.43, R) >droSec1.super_0 1829183 94 + 21120651 AACUGGGCGCGUUCGUUUUCACGCCGCUUGUCAGGUCGUUGAAAAUGUUACGACCGCCUAUCCUUCCUUCCAUCCUUGCAAACCUCCCCGAACA- ...(((((((((.((((((((((((........)).)).))))))))..)))..))))))..................................- ( -22.50, z-score = -1.50, R) >droYak2.chr3L 21862595 93 - 24197627 AACUGGGCGCGUUCGUUUUCACGCCGCUUGUCAGGUCGUUGAAAAUGUUACGAGCGCUUUUUCUUCCUUGCCAG-GACCAAACCACCCCGAACA- ...((((.(((((((((((((((((........)).)).))))).....)))))))).......((((....))-)).........))))....- ( -26.00, z-score = -0.82, R) >droEre2.scaffold_4784 9582216 90 + 25762168 AACUGGGCGCGUUCGUUUUCACGCCGCUUGUCAGGUCGUUGAAAAUGUUACGACCGCCUUUACCUCCUUGC----AAACCAACCACCCCGAACA- ....((((((((.((((((((((((........)).)).))))))))..)))..)))))............----...................- ( -21.10, z-score = -0.44, R) >droAna3.scaffold_13337 3840513 89 + 23293914 AACUGGGCGCGUUCGUUUUCACGCCGCUUGUCAGGUCGUUGAAAAUGUUACGACCUCCUUACCCGCGUCCCGACUUUCCCAGCCAAACU------ ..(((((.((((........))))(((..((.(((((((..........)))))))....))..)))..........))))).......------ ( -22.90, z-score = -0.97, R) >dp4.chrXR_group8 9177770 78 + 9212921 AACUGGGCGCGUUCGUUUUCACGCUGCCUGUCAGGUCGUUGAAAAUGUUACGACCUCCU---CCUCCCUUC----GAACACUCCA---------- ....((((((((........)))).))))...(((((((..........)))))))...---.........----..........---------- ( -19.10, z-score = -1.15, R) >droPer1.super_50 527452 78 - 557249 AACUGGGCGCGUUCGUUUUCACGCUGCCUGUCAGGUCGUUGAAAAUGUUACGACCUCCU---CCUCCCUUC----GAACACUCCA---------- ....((((((((........)))).))))...(((((((..........)))))))...---.........----..........---------- ( -19.10, z-score = -1.15, R) >droWil1.scaffold_180698 1449167 95 - 11422946 AACUGGGUGCGUUCAUUUUCACGCUGCUUGUCAGCUCGUUGAAAAUGUUACGACAAAAGGGGCGGCGUCCUCUUGGCCUACUCCAUUCUCGAAAC ...(((((((((.((((((((((..(((....))).)).))))))))..))).))...((((..(.(.((....))))..))))...)))).... ( -25.20, z-score = -0.61, R) >consensus AACUGGGCGCGUUCGUUUUCACGCCGCUUGUCAGGUCGUUGAAAAUGUUACGACCGCCUAUCCCUCCUUCC____GAACAAACCACCCCGAACA_ ....((((((((.((((((((((((........)).)).))))))))..)))..))))).................................... (-15.91 = -16.27 + 0.36)

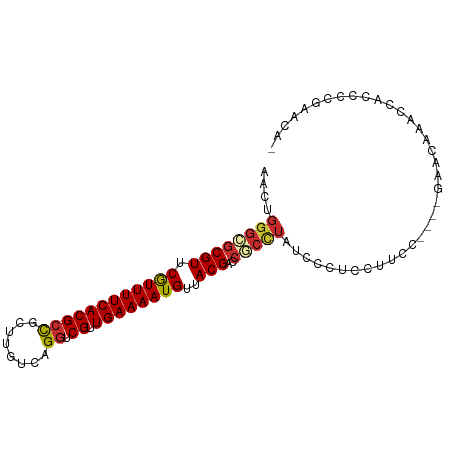

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:14:01 2011