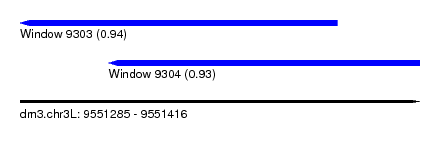
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,551,285 – 9,551,416 |
| Length | 131 |
| Max. P | 0.943652 |

| Location | 9,551,285 – 9,551,389 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.76 |
| Shannon entropy | 0.57656 |
| G+C content | 0.39265 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -9.93 |
| Energy contribution | -10.67 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
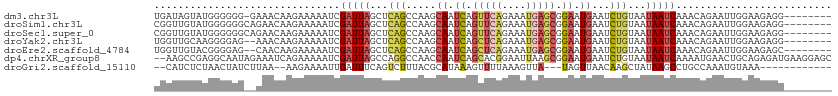
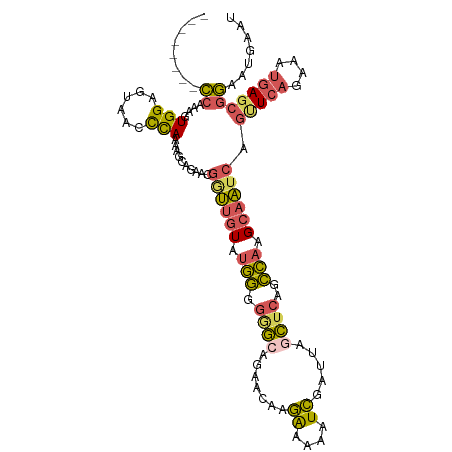
>dm3.chr3L 9551285 104 - 24543557 UGAUAGUAUGGGGGG-GAAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGUUCAGAAAUGAGCGGAAUGAAUCUGUAAUAAUCAAACAGAAUUGGAAGAGG-------- ((((.((.(((..((-(......((....))......)))..))).)).))))(((((....)))))........(((((.........)))))...........-------- ( -21.70, z-score = -2.09, R) >droSim1.chr3L 8935157 105 - 22553184 CGGUUGUAUGGGGGGCAGAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGUUCAGAAAUGAGCGGAAUGAAUCUGUAAUAAUCAAACAGAAUUGGAAGAGG-------- .((((((.(((.((((.((....((....))..)).))))..))).)))))).(((((....)))))........(((((.........)))))...........-------- ( -25.50, z-score = -2.66, R) >droSec1.super_0 1784847 105 - 21120651 CGGUUGUAUGGGGGGCAGAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGUUCAGAAAUGAGCGGAAUGAAUCUGUAAUAAUCAAACAGAAUUGGAAGAGG-------- .((((((.(((.((((.((....((....))..)).))))..))).)))))).(((((....)))))........(((((.........)))))...........-------- ( -25.50, z-score = -2.66, R) >droYak2.chr3L 21820404 103 + 24197627 UGGUUGCAAGGGGAG--AAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGCUCAGAAAUGAGCGGAAUGAAUCUGUAAUAAUCAAACAGAAUUGGAAGAGG-------- (((((((..((.(((--......((....))......)))..))..)))))))(((((....)))))........(((((.........)))))...........-------- ( -26.90, z-score = -3.27, R) >droEre2.scaffold_4784 9536329 103 - 25762168 UGGUUGUACGGGGAG--CAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGCUCAGAAAUGAGCGGAAUGAAUCUGUAAUAAUCAAACAGAAUUGGAAGAGC-------- ..(((((.......)--))))...............((((..((((.((.((.(((((....))))).)).))..(((((.........))))).))))..))))-------- ( -28.70, z-score = -3.46, R) >dp4.chrXR_group8 9135182 111 - 9212921 --AAGCCGAGGCAAUAGAAAUCAGAAAAAUCGAUUAGCCAGGCCAACCAAUCAGCACGGAAUUAAGCGGAAUGAAUCUGUAAUAAUCAAAAUGAACUGCAGAGAUGAAGGAGC --..(((..((((((.((...........)).))).))).)))...((..(((...((........))...))).((((((....((.....))..))))))......))... ( -18.60, z-score = -0.02, R) >droGri2.scaffold_15110 8841432 94 - 24565398 --CAUCUCUAACUAUCUUAA--AAGAAAAUUGAUUUCAGUCUUUACGCAUAAAGUUUUAAAGUUA---UAGUUAACAAGCUAUAAGCCUGCCAAAUGUAAA------------ --......(((((((.((((--((((((.....))))...(((((....)))))))))))....)---)))))).((.((.....)).))...........------------ ( -8.00, z-score = 1.13, R) >consensus UGGUUGUAUGGGGAG__AAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGUUCAGAAAUGAGCGGAAUGAAUCUGUAAUAAUCAAACAGAAUUGGAAGAGG________ ...............................(((((...(((.....((.((.(((((....))))).)).))...)))...))))).......................... ( -9.93 = -10.67 + 0.74)

| Location | 9,551,314 – 9,551,416 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.53 |
| Shannon entropy | 0.51417 |
| G+C content | 0.41649 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.48 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9551314 102 - 24543557 --------CCAAAGUGGAGUAACCCAAAAGCAGAAUGAUAGUAUGGGGGG-GAAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGUUCAGAAAUGAGCGGAAUGAAU --------((....(((......))).........((((.((.(((..((-(......((....))......)))..))).)).))))(((((....)))))))....... ( -21.90, z-score = -1.73, R) >droSim1.chr3L 8935186 103 - 22553184 --------CCAAAGUGGAGUAACCCAAAAGCAGAACGGUUGUAUGGGGGGCAGAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGUUCAGAAAUGAGCGGAAUGAAU --------((....(((......)))..........((((((.(((.((((.((....((....))..)).))))..))).)))))).(((((....)))))))....... ( -25.40, z-score = -2.15, R) >droSec1.super_0 1784876 103 - 21120651 --------CCAAAGUGGAGUAACCCAAAAGCAGAACGGUUGUAUGGGGGGCAGAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGUUCAGAAAUGAGCGGAAUGAAU --------((....(((......)))..........((((((.(((.((((.((....((....))..)).))))..))).)))))).(((((....)))))))....... ( -25.40, z-score = -2.15, R) >droEre2.scaffold_4784 9536358 101 - 25762168 --------CCAAAGUGGUGCAACCCAAACGCGGAAUGGUUGUACGGGGAGC--AACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGCUCAGAAAUGAGCGGAAUGAAU --------((...((.((((((((............))))))))((.((((--.....((....)).....))))..))..)).....(((((....)))))))....... ( -29.60, z-score = -2.45, R) >droGri2.scaffold_15110 8841457 106 - 24565398 AUAAAGCUGAACAGUGGGGCCUUUUAUAAUCUUACCACAUCUCUAACUAUCUUA--AAAGAAAAUUGAUUUCAGUCUUUACGCAUAAAGUUUUAAAGUUA---UAGUUAAC .((((((((((..((((.................))))...........(((..--..)))........))))).)))))................((((---....)))) ( -12.93, z-score = 0.70, R) >consensus ________CCAAAGUGGAGUAACCCAAAAGCAGAACGGUUGUAUGGGGGGCAGAACAAGAAAAAUCGAUUAGCUCAGCCAAGCAAUCAGUUCAGAAAUGAGCGGAAUGAAU ........((....(((......)))..........((((((.(((.((((.......((....)).....))))..))).)))))).(((((....)))))))....... (-13.44 = -13.48 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:55 2011