| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,534,181 – 9,534,271 |
| Length | 90 |
| Max. P | 0.922332 |

| Location | 9,534,181 – 9,534,271 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 65.60 |
| Shannon entropy | 0.53825 |
| G+C content | 0.52995 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -19.77 |
| Energy contribution | -21.28 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
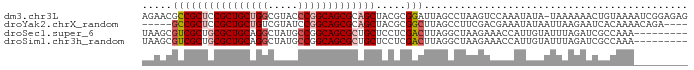
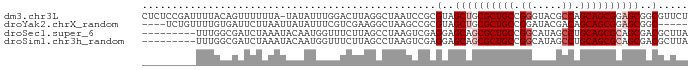
>dm3.chr3L 9534181 90 + 24543557 AGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUUAGCCUAAGUCCAAAUAUA-UAAAAAACUGUAAAAUCGGAGAG .....((.(((.(((((((((.....))))))))).)))...))(((((......))))).......-.......(((......))).... ( -28.40, z-score = -1.56, R) >droYak2.chrX_random 1263068 82 - 1802292 -----GCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGCUUAGCCUUCGACGAAAUAUAAUUAAGAAUCACAAAACAGA---- -----(((((...((((((((((....)))))..)))))...)))))........................................---- ( -23.30, z-score = -0.88, R) >droSec1.super_6 695964 82 - 4358794 UAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAGAAACCAUUGUAUUUAGAUCGCCAAA--------- ...(((((((.(((((((.(((...))).))))))).))...........((........))..........)).)))....--------- ( -27.50, z-score = -0.68, R) >droSim1.chr3h_random 1352278 82 + 1452968 UAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAGAAACCAUUGUAUUUAGAUCGCCAAA--------- ...(((((((.(((((((.(((...))).))))))).))...........((........))..........)).)))....--------- ( -27.50, z-score = -0.68, R) >consensus UAAGCGCCGCUCCGCUGCAGGCGAAGCCGGCAGCGCAGCUACGCGACUUAGCCUAAGAAAAAAUAGAAUUAAGAACGCCAAA_________ .....((((((((((((((((.....))))))))))))).....)))............................................ (-19.77 = -21.28 + 1.50)
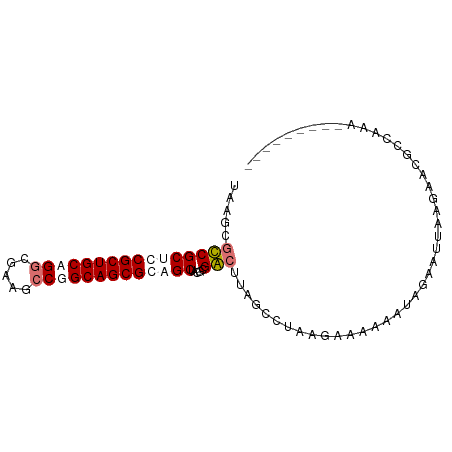
| Location | 9,534,181 – 9,534,271 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 65.60 |
| Shannon entropy | 0.53825 |
| G+C content | 0.52995 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -19.15 |
| Energy contribution | -20.40 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9534181 90 - 24543557 CUCUCCGAUUUUACAGUUUUUUA-UAUAUUUGGACUUAGGCUAAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCU ...(((((...............-.....)))))....((.....))((((.(((.((((((.((.....)).)))))).))).))))... ( -27.25, z-score = -0.18, R) >droYak2.chrX_random 1263068 82 + 1802292 ----UCUGUUUUGUGAUUCUUAAUUAUAUUUCGUCGAAGGCUAAGCCGCGUAGCUGCGCUGCCGGAUACGACAGCAGCGGAGCGGC----- ----...(((((.((((...............)))))))))...(((((....((((((((.((....)).)))).)))).)))))----- ( -25.06, z-score = 0.09, R) >droSec1.super_6 695964 82 + 4358794 ---------UUUGGCGAUCUAAAUACAAUGGUUUCUUAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUA ---------...((((.((...........((((((..((....))..)))))).(((((((.(((...))).)))))))...)))))).. ( -32.80, z-score = -2.03, R) >droSim1.chr3h_random 1352278 82 - 1452968 ---------UUUGGCGAUCUAAAUACAAUGGUUUCUUAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUA ---------...((((.((...........((((((..((....))..)))))).(((((((.(((...))).)))))))...)))))).. ( -32.80, z-score = -2.03, R) >consensus _________UUUGGCGAUCUAAAUAAAAUGGUGUCUUAGCCUAAGCCGAGGAGCAGCGCUGCCGGCAAAGCCAGCAGCGCAGCGACGCUUA .................................................(..((((((((((.((.....)).))))))))))..)..... (-19.15 = -20.40 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:53 2011