| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,490,627 – 9,490,725 |
| Length | 98 |
| Max. P | 0.936621 |

| Location | 9,490,627 – 9,490,725 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.57 |
| Shannon entropy | 0.42630 |
| G+C content | 0.50729 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.77 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
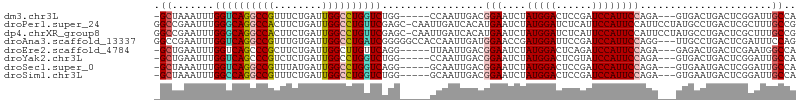
>dm3.chr3L 9490627 98 + 24543557 -GCUAAAUUUGGUCAGGCCGUUUCUGAUUGGCCUGGUCUGG-----CCAAUUGACGGAAUCUAUGGACUCCGAUCCAUUCCAGA---GUGACUGACUCGGAUUGCCA -((..(((((((((((.(((((...((((((((......))-----)))))))))))..((((((((......)))))...)))---....))))).)))))))).. ( -34.40, z-score = -2.05, R) >droPer1.super_24 886103 106 - 1556852 GGCCGAAUUUGGGCAGGCCACUUCUGAUUGGCCUGUUCGAGC-CAAUUGAUCACAUGAAUCUAUGGAUCUCAUUCCAUUCCAUUCCUAUGCCUGACUCGCUUUGCCG (((....(((((((((((((........))))))))))))).-.....((((((((((((..(((((......)))))...))))..)))..))).)).....))). ( -32.10, z-score = -1.99, R) >dp4.chrXR_group8 2384127 106 + 9212921 GGCCGAAUUUGGGCAGGCCACUUCUGAUUGGCCUGUUCGAGC-CAAUUGAUCACAUGAAUCUAUGGAUCUCAUUCCAUUCCAUUCCUAUGCCUGACUCGCUUUGCCG (((....(((((((((((((........))))))))))))).-.....((((((((((((..(((((......)))))...))))..)))..))).)).....))). ( -32.10, z-score = -1.99, R) >droAna3.scaffold_13337 3746218 104 + 23293914 GGCCGAAUUUGGUCAGGCCGUUUGUGAUUGGCCUGAUCGGGGGCCACCAAUUGAUGGAACCGAUGGAUUCCGAUCCAUUCCAGG---UUGCCUGACUCGAUUUCCAG ((((...(((((((((((((........)))))))))))))))))...((((((.(((((((((((((....)))))))...))---)).))....))))))..... ( -41.30, z-score = -1.90, R) >droEre2.scaffold_4784 9482659 98 + 25762168 -GCUGAAUUUGGUCAGCCCGCUUCUGAUUGGCUUGUUCAGG-----UUAAUUGACGGAAUCUAUGGACUCAGAUCCAUUCCAGA---GAGACUGACUCGAAUGGCCA -(((..((((((((((..(..(((((((((((((....)))-----))))))...(((....(((((......)))))))))))---).).))))).)))))))).. ( -26.10, z-score = 0.06, R) >droYak2.chr3L 21764176 98 - 24197627 -GCUGAAUUUGGUCAGCCCGUCUCUGAUUGGCCUGGUCUGG-----CCAAUUGACGGAAUCUAUGGACUCGUAUCCAUUCCAGA---GUGACUGACUCGGAUUGCCA -(((((......)))))(((((...((((((((......))-----)))))))))))((((((((((......)))))....((---((.....))))))))).... ( -35.80, z-score = -2.41, R) >droSec1.super_0 1731698 98 + 21120651 -GCUAAAUUUGGUCAGGCCGUUUAUGAUUGGCCUGGUCAGG-----GCAAUUGACGGAAUCUAUGGACUCCGAUCCAUUCCAGA---GUGAAUGACUCGGAUUGCCA -.......(((..(((((((........)))))))..)))(-----((((((...(((....(((((......)))))))).((---((.....)))).))))))). ( -38.60, z-score = -3.18, R) >droSim1.chr3L 8880442 98 + 22553184 -GCUAAAUUUGGCCAGGCCGUUUCUGAUUGGCCUGGUCUGG-----GCAAUUGACGGAAUCUAUGGACUCCGAUCCAUUCCAGA---GUGAAUGACUCGGAUUGCCA -.........((((((((((........))))))))))..(-----((((((...(((....(((((......)))))))).((---((.....)))).))))))). ( -39.80, z-score = -3.38, R) >consensus _GCUGAAUUUGGUCAGGCCGUUUCUGAUUGGCCUGGUCGGG_____CCAAUUGACGGAAUCUAUGGACUCCGAUCCAUUCCAGA___GUGACUGACUCGGAUUGCCA .((.......((((((((((........)))))))))).................(((....(((((......))))))))......................)).. (-19.76 = -19.77 + 0.02)
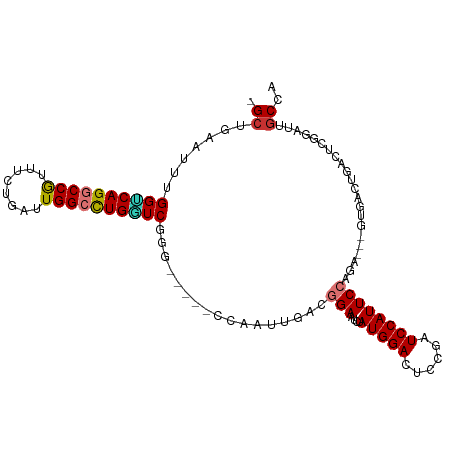
| Location | 9,490,627 – 9,490,725 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Shannon entropy | 0.42630 |
| G+C content | 0.50729 |
| Mean single sequence MFE | -33.51 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9490627 98 - 24543557 UGGCAAUCCGAGUCAGUCAC---UCUGGAAUGGAUCGGAGUCCAUAGAUUCCGUCAAUUGG-----CCAGACCAGGCCAAUCAGAAACGGCCUGACCAAAUUUAGC- ((((.(((((..((((....---.))))..)))))(((((((....))))))).......)-----)))...((((((..........))))))............- ( -33.60, z-score = -2.65, R) >droPer1.super_24 886103 106 + 1556852 CGGCAAAGCGAGUCAGGCAUAGGAAUGGAAUGGAAUGAGAUCCAUAGAUUCAUGUGAUCAAUUG-GCUCGAACAGGCCAAUCAGAAGUGGCCUGCCCAAAUUCGGCC .(((....((((((((.((((.((((...(((((......)))))..)))).)))).....)))-)))))..(((((((........)))))))..........))) ( -36.30, z-score = -2.32, R) >dp4.chrXR_group8 2384127 106 - 9212921 CGGCAAAGCGAGUCAGGCAUAGGAAUGGAAUGGAAUGAGAUCCAUAGAUUCAUGUGAUCAAUUG-GCUCGAACAGGCCAAUCAGAAGUGGCCUGCCCAAAUUCGGCC .(((....((((((((.((((.((((...(((((......)))))..)))).)))).....)))-)))))..(((((((........)))))))..........))) ( -36.30, z-score = -2.32, R) >droAna3.scaffold_13337 3746218 104 - 23293914 CUGGAAAUCGAGUCAGGCAA---CCUGGAAUGGAUCGGAAUCCAUCGGUUCCAUCAAUUGGUGGCCCCCGAUCAGGCCAAUCACAAACGGCCUGACCAAAUUCGGCC .......((((((((((...---))))...(((.((((...(((((((((.....)))))))))...))))(((((((..........)))))))))).)))))).. ( -35.50, z-score = -1.02, R) >droEre2.scaffold_4784 9482659 98 - 25762168 UGGCCAUUCGAGUCAGUCUC---UCUGGAAUGGAUCUGAGUCCAUAGAUUCCGUCAAUUAA-----CCUGAACAAGCCAAUCAGAAGCGGGCUGACCAAAUUCAGC- .........(.(((((((((---(((((.((((((....))))))........(((.....-----..))).........)))).)).))))))))).........- ( -21.50, z-score = 0.66, R) >droYak2.chr3L 21764176 98 + 24197627 UGGCAAUCCGAGUCAGUCAC---UCUGGAAUGGAUACGAGUCCAUAGAUUCCGUCAAUUGG-----CCAGACCAGGCCAAUCAGAGACGGGCUGACCAAAUUCAGC- ......(((((((.....))---)).)))((((((....)))))).....(((((.(((((-----((......)))))))....)))))(((((......)))))- ( -34.50, z-score = -2.76, R) >droSec1.super_0 1731698 98 - 21120651 UGGCAAUCCGAGUCAUUCAC---UCUGGAAUGGAUCGGAGUCCAUAGAUUCCGUCAAUUGC-----CCUGACCAGGCCAAUCAUAAACGGCCUGACCAAAUUUAGC- .((((((((((((.....))---)).)))...((.(((((((....)))))))))..))))-----).((..((((((..........))))))..))........- ( -33.60, z-score = -3.24, R) >droSim1.chr3L 8880442 98 - 22553184 UGGCAAUCCGAGUCAUUCAC---UCUGGAAUGGAUCGGAGUCCAUAGAUUCCGUCAAUUGC-----CCAGACCAGGCCAAUCAGAAACGGCCUGGCCAAAUUUAGC- .((((((((((((.....))---)).)))...((.(((((((....)))))))))..))))-----)..(.(((((((..........))))))).).........- ( -36.80, z-score = -3.63, R) >consensus UGGCAAUCCGAGUCAGUCAC___UCUGGAAUGGAUCGGAGUCCAUAGAUUCCGUCAAUUGA_____CCAGACCAGGCCAAUCAGAAACGGCCUGACCAAAUUCAGC_ .((......(((((...((......))..((((((....)))))).))))).....................((((((..........)))))).)).......... (-16.69 = -17.38 + 0.69)

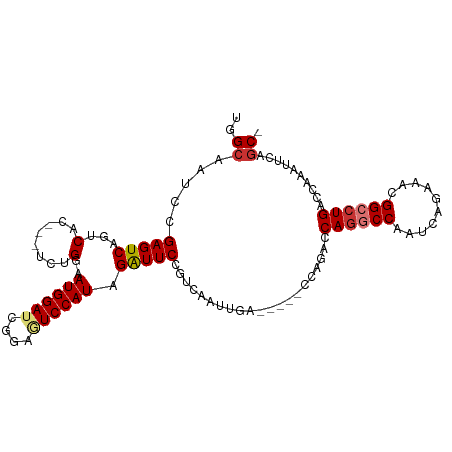
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:50 2011