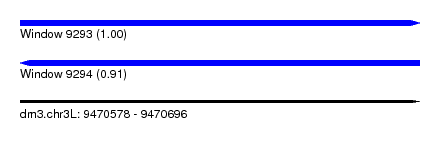
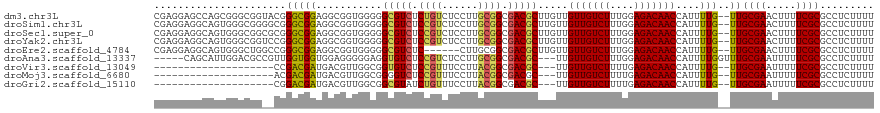
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,470,578 – 9,470,696 |
| Length | 118 |
| Max. P | 0.995994 |

| Location | 9,470,578 – 9,470,696 |
|---|---|
| Length | 118 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.81 |
| Shannon entropy | 0.39239 |
| G+C content | 0.56385 |
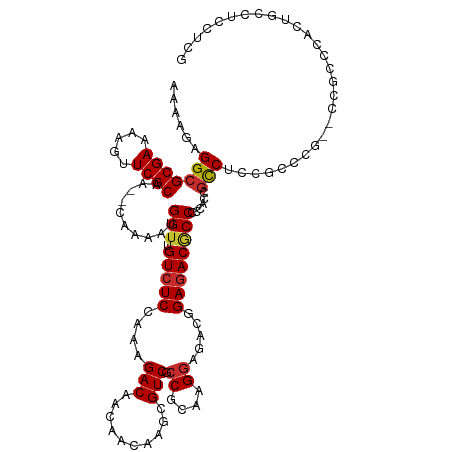
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -22.47 |
| Energy contribution | -23.21 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
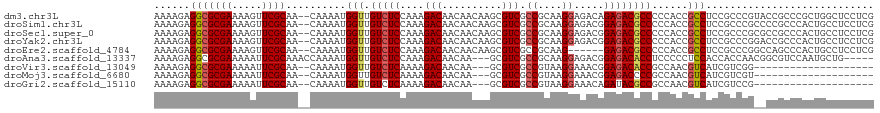
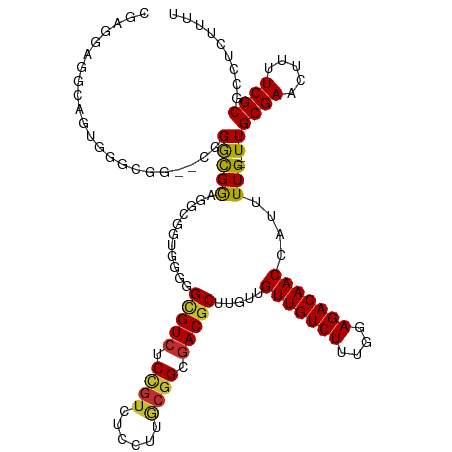
>dm3.chr3L 9470578 118 + 24543557 AAAAGAGGCGCGAAAAGUUCGCAA--CAAAAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAGGAGACAGAGACGCCCCCACCGCCUCCGCCCGUACCGCCCGCUGGCUCCUCG ....(((((((((.....))))..--.......(((((((....))))))).....(((((.((....)).......))))).......))))).(((.((.......)).)))...... ( -38.20, z-score = -2.44, R) >droSim1.chr3L 8859017 118 + 22553184 AAAAGAGGCGCGAAAAGUUCGCAA--CAAAAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAGGAGACGGAGACGCCCCCACCGCCUCCGCCCGCCCCGCCCACUGCCUCCUCG ....(((((((((.....))))..--.......(((((((....))))))).....(((((.((....)).)))((((.((.......)).))))))..............))))).... ( -40.00, z-score = -3.36, R) >droSec1.super_0 1711691 118 + 21120651 AAAAGAGGCGCGAAAAGUUCGCAA--CAAAAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAGGAGACGGAGACGCCCCCACCGCCUCCGCCCGCGCCGCCCACUGCCUCCUCG ....(((((((((.....))))..--.......(((((((....))))))).....(((((.((....)).)))((((.((.......)).))))....))..........))))).... ( -40.90, z-score = -2.99, R) >droYak2.chr3L 21744001 118 - 24197627 AAAAGAGGCGCGAAAAGUUCGCAA--CAAAAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAGGAGACGGAGACGCCCCCACCGCCUCCGCCCGGACCGCCCACUGCCUCCUCG ....(((((((((.....))))..--.......(((((((....))))))).....(((((.((....)).)))((((.((.......)).))))))..((.....))...))))).... ( -40.80, z-score = -2.99, R) >droEre2.scaffold_4784 9462259 112 + 25762168 AAAAGAGGCGCGAAAAGUUCGCAA--CAAAAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAG------GAGACGCCCCCACCGCCUCCGCCCGGCCAGCCCACUGCCUCCUCG ....(((((((((.....))))..--.......(((((((....))))))).....(((((.((....)------).))))).......))))).....(((.((....)))))...... ( -40.80, z-score = -4.34, R) >droAna3.scaffold_13337 3724097 112 + 23293914 AAAAGAGGCGCGAAAAAUUCGCAAACCAAAAUGGUUGUCUCCAAAGACAACAA---GCGUCGCCGCAAGGAGACGGAGACACCUCCCCCUCCACCACCAACGGCGUCCAAUGCUG----- ....(((((((((.....))))...........(((((((....)))))))..---))(((.((....)).)))((((....))))..))).........((((((...))))))----- ( -36.00, z-score = -3.46, R) >droVir3.scaffold_13049 16958454 95 - 25233164 AAAAGAGGCGCGAAAAAUUCGCAA--CAAAAUGGUUGUCUCAAAAGACAACAA---GCGUCGCCGUAAGGAAACGGAGACACCGCCAACGUCAUCGUCGG-------------------- ......(((((((.....))))..--.......(((((((....)))))))..---..(((.((((......)))).)))...))).(((....)))...-------------------- ( -28.80, z-score = -2.53, R) >droMoj3.scaffold_6680 12829011 95 - 24764193 AAAAGAGGCGCGAAAAAUUCGCAA--CAAAAUGGUUGUCUCAAAAGACAACAA---GCGUCGCCGUAAGGAAACGGAGACCCCGCCAACGUCAUCGUCGU-------------------- ......(((((((.....))))..--.......(((((((....)))))))..---..(((.((((......)))).)))...))).(((....)))...-------------------- ( -27.90, z-score = -2.17, R) >droGri2.scaffold_15110 17257135 95 - 24565398 AAAAGAGGCGCGAAAAAUUCGCAA--CAAAAUGGUUGUCUCAAAAGACAACAA---GCGUCGCCGUAAGGAAACAGAUACGCCGCCAACGUCAUCGUCCG-------------------- ....((.((((((.....))))..--.......(((((((....)))))))..---)).)).......(((....((((((.......))).))).))).-------------------- ( -25.70, z-score = -2.10, R) >consensus AAAAGAGGCGCGAAAAGUUCGCAA__CAAAAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAGGAGACGGAGACGCCCCCACCGCCUCCGCCCG__CCGCCCACUGCCUCCUCG ......(((((((.....))))..........(((.(((((....(((..........))).((....)).....))))))))......)))............................ (-22.47 = -23.21 + 0.74)
| Location | 9,470,578 – 9,470,696 |
|---|---|
| Length | 118 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.81 |
| Shannon entropy | 0.39239 |
| G+C content | 0.56385 |
| Mean single sequence MFE | -44.31 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.06 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 9470578 118 - 24543557 CGAGGAGCCAGCGGGCGGUACGGGCGGAGGCGGUGGGGGCGUCUCUGUCUCCUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUUUUG--UUGCGAACUUUUCGCGCCUCUUUU .(((((((.((((.((.(((.(((((((((((.......)))))))))))...))).))..)))).)))(((((((....))))))).......--..((((.....)))).)))).... ( -47.70, z-score = -0.94, R) >droSim1.chr3L 8859017 118 - 22553184 CGAGGAGGCAGUGGGCGGGGCGGGCGGAGGCGGUGGGGGCGUCUCCGUCUCCUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUUUUG--UUGCGAACUUUUCGCGCCUCUUUU .(((((((((((((((((((.(((((((((((.......)))))))))))))))))(((((((.....))))))).(((....))))))))...--..((((.....)))))))))))). ( -58.90, z-score = -3.68, R) >droSec1.super_0 1711691 118 - 21120651 CGAGGAGGCAGUGGGCGGCGCGGGCGGAGGCGGUGGGGGCGUCUCCGUCUCCUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUUUUG--UUGCGAACUUUUCGCGCCUCUUUU .(((((((((((((((((((((((((..(.((..(((((((....)))))))...)).)..)))))).))))))..(((....))))))))...--..((((.....)))))))))))). ( -54.70, z-score = -2.32, R) >droYak2.chr3L 21744001 118 + 24197627 CGAGGAGGCAGUGGGCGGUCCGGGCGGAGGCGGUGGGGGCGUCUCCGUCUCCUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUUUUG--UUGCGAACUUUUCGCGCCUCUUUU .((((((((((..((((..(((.(.((((((((.((.....)).)))))))).).)))...))))..))(((((((....))))))).......--..((((.....)))))))))))). ( -54.20, z-score = -2.41, R) >droEre2.scaffold_4784 9462259 112 - 25762168 CGAGGAGGCAGUGGGCUGGCCGGGCGGAGGCGGUGGGGGCGUCUC------CUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUUUUG--UUGCGAACUUUUCGCGCCUCUUUU .((((((((((..((((.((((.(.(((((((.......))))))------).).)))).).)))..))(((((((....))))))).......--..((((.....)))))))))))). ( -51.40, z-score = -2.57, R) >droAna3.scaffold_13337 3724097 112 - 23293914 -----CAGCAUUGGACGCCGUUGGUGGUGGAGGGGGAGGUGUCUCCGUCUCCUUGCGGCGACGC---UUGUUGUCUUUGGAGACAACCAUUUUGGUUUGCGAAUUUUUCGCGCCUCUUUU -----..((.......((((..(((((((((((.((((....)))).)))))..(((....)))---....(((((....))))))))))).))))..((((.....))))))....... ( -40.20, z-score = -1.13, R) >droVir3.scaffold_13049 16958454 95 + 25233164 --------------------CCGACGAUGACGUUGGCGGUGUCUCCGUUUCCUUACGGCGACGC---UUGUUGUCUUUUGAGACAACCAUUUUG--UUGCGAAUUUUUCGCGCCUCUUUU --------------------..((((....))))(((((((((.((((......)))).)))))---).(((((((....))))))).......--..((((.....)))))))...... ( -32.80, z-score = -3.41, R) >droMoj3.scaffold_6680 12829011 95 + 24764193 --------------------ACGACGAUGACGUUGGCGGGGUCUCCGUUUCCUUACGGCGACGC---UUGUUGUCUUUUGAGACAACCAUUUUG--UUGCGAAUUUUUCGCGCCUCUUUU --------------------..((((....))))(((((.(((.((((......)))).))).)---).(((((((....))))))).......--..((((.....)))))))...... ( -30.60, z-score = -2.15, R) >droGri2.scaffold_15110 17257135 95 + 24565398 --------------------CGGACGAUGACGUUGGCGGCGUAUCUGUUUCCUUACGGCGACGC---UUGUUGUCUUUUGAGACAACCAUUUUG--UUGCGAAUUUUUCGCGCCUCUUUU --------------------..((((....))))((((((((..((((......))))..))))---).(((((((....))))))).......--..((((.....)))))))...... ( -28.30, z-score = -1.60, R) >consensus CGAGGAGGCAGUGGGCGG__CGGGCGGAGGCGGUGGGGGCGUCUCCGUCUCCUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUUUUG__UUGCGAACUUUUCGCGCCUCUUUU ..................................((..(((((.((((......)))).))))).....(((((((....)))))))...........((((.....)))).))...... (-23.98 = -24.06 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:47 2011