| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,450,948 – 9,451,039 |
| Length | 91 |
| Max. P | 0.864769 |

| Location | 9,450,948 – 9,451,039 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 64.54 |
| Shannon entropy | 0.64666 |
| G+C content | 0.50031 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -10.79 |
| Energy contribution | -12.44 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
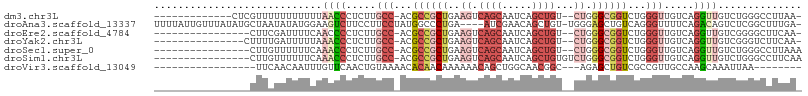
>dm3.chr3L 9450948 91 + 24543557 -------------CUCGUUUUUUUUUUUAACCCUCUUGCC-ACGCCGCUGAAGUCAGCAAUCAGCUGU--CUGGGCGGUCUGGGUUGUCAGGUUGUCUGGGCCUUAA- -------------..............(((((.....(((-..((((((..((.((((.....)))).--)).))))))...))).....)))))............- ( -26.80, z-score = -0.78, R) >droAna3.scaffold_13337 3703233 102 + 23293914 UUUUAUUGUUUAUAUGCUAAUAUAUGGAAGUCUUCCUUCCUAUGGCCCUGA----AUCGAACAGCUGU-UGGGAGCUGUCAGGGUUUUCAGACAGUCUCGGCUUUGA- ....(((((((.........((((.(((((.....)))))))))(((((((----......(((((..-....))))))))))))....)))))))...........- ( -26.10, z-score = -0.48, R) >droEre2.scaffold_4784 9442403 88 + 25762168 ----------------CUUCGAUUUUCAACCCCUCUUGCC-ACGCCGCUGAAGUCAGCAAUCAGCUGU--CUGGGCGGUCUGGGUUGUCAGGUUGUCGGGGCUUCAA- ----------------((((((....(((((...(...((-(.((((((..((.((((.....)))).--)).)))))).)))...)...)))))))))))......- ( -32.50, z-score = -1.78, R) >droYak2.chr3L 21724231 89 - 24197627 ---------------CUUUUGAUUUUUAAACCCUCUUGCC-ACGCCGCUGAAGUCAGCAAUCAGCUGU--CUGGGCGGUCUGGGUUGUCAGGUUGUCGGGUCUUCAA- ---------------.....(((((...((((.....(((-..((((((..((.((((.....)))).--)).))))))...))).....))))...))))).....- ( -29.20, z-score = -1.72, R) >droSec1.super_0 1692113 89 + 21120651 ----------------CUUGUUUUUUCAAACCCUCUUGCC-ACGCCGCUGAAGUCAGCAAUCAGCUGU--CUGGGCGGUCUGGGUUGUCAGGUUGUCUGGGCCUUAAA ----------------...((((.....((((.....(((-..((((((..((.((((.....)))).--)).))))))...))).....))))....))))...... ( -27.30, z-score = -0.63, R) >droSim1.chr3L 8841782 91 + 22553184 ----------------CUUGUUUUUUCAAACCCUCUUGCC-ACGCCGCUGAAGUCAGCAAUCAGCUGUGUCUGGGCGGUCUGGGUUGUCAGGUUGUCUGGGCCUUCAA ----------------............(((((..(((((-((((.(((((.........))))).))))...)))))...)))))...(((((.....))))).... ( -25.40, z-score = 0.33, R) >droVir3.scaffold_13049 16933130 80 - 25233164 -----------------UUCAACAAUUUGUUCAACUGUAAAACACAACAAAAAACAGCUGGCAACGGC---AGAGCUGUCGCCGUUGCCAAGCAAAUUAA-------- -----------------......((((((((...((((...............)))).((((((((((---.((....))))))))))))))))))))..-------- ( -24.76, z-score = -3.67, R) >consensus ________________CUUCUAUUUUCAAACCCUCUUGCC_ACGCCGCUGAAGUCAGCAAUCAGCUGU__CUGGGCGGUCUGGGUUGUCAGGUUGUCUGGGCCUUAA_ ............................((((.....(((...((((((.....((((.....))))......))))))...))).....)))).............. (-10.79 = -12.44 + 1.66)


| Location | 9,450,948 – 9,451,039 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 64.54 |
| Shannon entropy | 0.64666 |
| G+C content | 0.50031 |
| Mean single sequence MFE | -24.49 |
| Consensus MFE | -9.58 |
| Energy contribution | -10.18 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
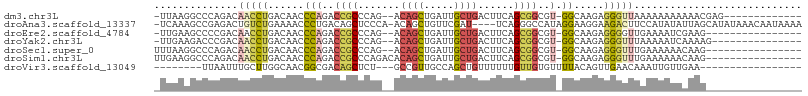
>dm3.chr3L 9450948 91 - 24543557 -UUAAGGCCCAGACAACCUGACAACCCAGACCGCCCAG--ACAGCUGAUUGCUGACUUCAGCGGCGU-GGCAAGAGGGUUAAAAAAAAAAACGAG------------- -....(((((.......(((......))).((((((((--....)))...((((....)))))))).-)......)))))...............------------- ( -23.10, z-score = -1.25, R) >droAna3.scaffold_13337 3703233 102 - 23293914 -UCAAAGCCGAGACUGUCUGAAAACCCUGACAGCUCCCA-ACAGCUGUUCGAU----UCAGGGCCAUAGGAAGGAAGACUUCCAUAUAUUAGCAUAUAAACAAUAAAA -.............((.((((...(((((((((((....-..)))))......----)))))).....(((((.....))))).....)))))).............. ( -23.30, z-score = -1.78, R) >droEre2.scaffold_4784 9442403 88 - 25762168 -UUGAAGCCCCGACAACCUGACAACCCAGACCGCCCAG--ACAGCUGAUUGCUGACUUCAGCGGCGU-GGCAAGAGGGGUUGAAAAUCGAAG---------------- -....((((((......(((......))).((((((((--....)))...((((....)))))))).-)......))))))...........---------------- ( -26.50, z-score = -1.56, R) >droYak2.chr3L 21724231 89 + 24197627 -UUGAAGACCCGACAACCUGACAACCCAGACCGCCCAG--ACAGCUGAUUGCUGACUUCAGCGGCGU-GGCAAGAGGGUUUAAAAAUCAAAAG--------------- -((((((((((......(((......))).((((((((--....)))...((((....)))))))).-)......)))))).....))))...--------------- ( -24.40, z-score = -2.08, R) >droSec1.super_0 1692113 89 - 21120651 UUUAAGGCCCAGACAACCUGACAACCCAGACCGCCCAG--ACAGCUGAUUGCUGACUUCAGCGGCGU-GGCAAGAGGGUUUGAAAAAACAAG---------------- ....((((((.......(((......))).((((((((--....)))...((((....)))))))).-)......))))))...........---------------- ( -23.70, z-score = -0.97, R) >droSim1.chr3L 8841782 91 - 22553184 UUGAAGGCCCAGACAACCUGACAACCCAGACCGCCCAGACACAGCUGAUUGCUGACUUCAGCGGCGU-GGCAAGAGGGUUUGAAAAAACAAG---------------- ....((((((.......(((......))).((((((((......)))...((((....)))))))).-)......))))))...........---------------- ( -23.90, z-score = -0.83, R) >droVir3.scaffold_13049 16933130 80 + 25233164 --------UUAAUUUGCUUGGCAACGGCGACAGCUCU---GCCGUUGCCAGCUGUUUUUUGUUGUGUUUUACAGUUGAACAAAUUGUUGAA----------------- --------.(((((((((.((((((((((.......)---)))))))))))).((((.((((........))))..)))))))))).....----------------- ( -26.50, z-score = -2.71, R) >consensus _UUAAAGCCCAGACAACCUGACAACCCAGACCGCCCAG__ACAGCUGAUUGCUGACUUCAGCGGCGU_GGCAAGAGGGUUUAAAAAUAAAAG________________ ..............(((((.....((....((((.......((((.....))))......))))....)).....)))))............................ ( -9.58 = -10.18 + 0.60)

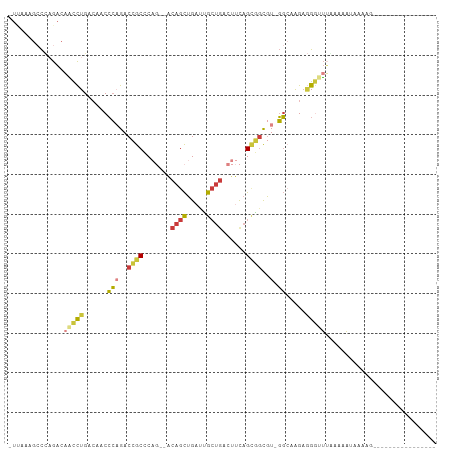
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:45 2011