| Sequence ID | dm3.chr3L |
|---|---|
| Location | 9,407,342 – 9,407,398 |
| Length | 56 |
| Max. P | 0.809069 |

| Location | 9,407,342 – 9,407,398 |
|---|---|
| Length | 56 |
| Sequences | 14 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 81.86 |
| Shannon entropy | 0.40840 |
| G+C content | 0.39039 |
| Mean single sequence MFE | -11.22 |
| Consensus MFE | -7.36 |
| Energy contribution | -7.16 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
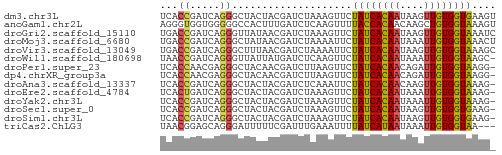
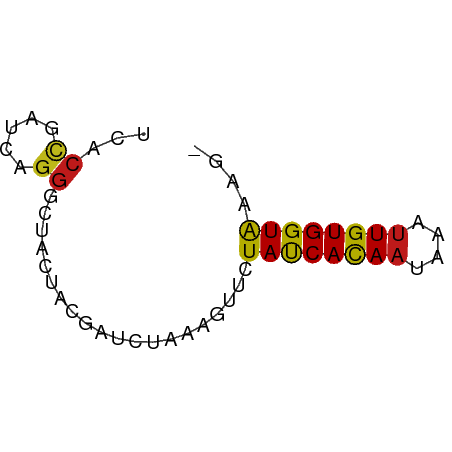
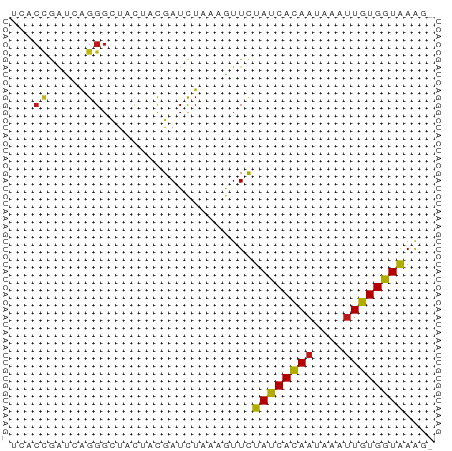
>dm3.chr3L 9407342 56 - 24543557 UCACCGAUCAGGGCUACUACGAUCUAAAGUUCUAUCACAAUAAGUUGUGGUGAAGU ((((((((.((((((............)))))))))((((....)))))))))... ( -13.60, z-score = -1.72, R) >anoGam1.chr2L 21499622 56 - 48795086 AGGGUGGUGGGGGCCACUUUGAUCUCAAGUUUUACCACAACAAGCUGUGGUAAAGU ((((((((....)))))))).........((((((((((......)))))))))). ( -19.20, z-score = -1.98, R) >droGri2.scaffold_15110 15576137 56 + 24565398 UGACCGAUCAGGGUUAUAACGAUCUAAAGUUCUAUCACAAUAAGUUGUGGUAAAUC (((((......)))))................((((((((....)))))))).... ( -11.00, z-score = -1.44, R) >droMoj3.scaffold_6680 14231336 56 + 24764193 UGACCGAUCAGGGCUAUAACGAUCUAAAAUUCUAUCACAAUAAAUUGUGGUAAACU .....((((.(........)))))........((((((((....)))))))).... ( -9.40, z-score = -1.75, R) >droVir3.scaffold_13049 10255413 56 - 25233164 UGACCGAUCAGGGCUUUAACGAUCUAAAAUUCUAUCACAAUAAGUUGUGGUAAAGC ...((.....))((((((..((........))..((((((....)))))))))))) ( -9.80, z-score = -1.02, R) >droWil1.scaffold_180698 1571636 55 - 11422946 UAACCGAUCAGGGUUAUUAUGAUCUCAAGUUCUAUCACAAUAAAUUGUGGUAAGC- .....(((((.........)))))........((((((((....))))))))...- ( -10.80, z-score = -1.80, R) >droPer1.super_23 1443233 55 - 1662726 UCACCAACGAGGGCUACAACGAUCUUAAGUUCUAUCACAACAGAUUGUGGUAAGG- ...((.....(((((............)))))((((((((....)))))))).))- ( -9.30, z-score = -0.59, R) >dp4.chrXR_group3a 1413607 55 - 1468910 UCACCAACGAGGGCUACAACGAUCUUAAGUUCUAUCACAACAGAUUGUGGUAAGG- ...((.....(((((............)))))((((((((....)))))))).))- ( -9.30, z-score = -0.59, R) >droAna3.scaffold_13337 16993834 55 - 23293914 UCACCGAUCAGGGCUACUACGAUCUCAAAUUCUAUCACAACAAGUUGUGGUAAAG- .....((((.((....))..))))........((((((((....))))))))...- ( -11.10, z-score = -2.12, R) >droEre2.scaffold_4784 9402246 55 - 25762168 UCACUGAUCAGGGCUACUACGAUCUAAAGUUCUAUCACAAUAAAUUGUGGUAAAG- .....((((.((....))..))))........((((((((....))))))))...- ( -9.60, z-score = -1.56, R) >droYak2.chr3L 21684264 55 + 24197627 UCACCGAUCAGGGCUACUACGAUCUAAAGUUCUAUCACAAUAAAUUGUGGUAAAG- .....((((.((....))..))))........((((((((....))))))))...- ( -9.90, z-score = -1.65, R) >droSec1.super_0 1652476 55 - 21120651 UCACCGAUCAGGGCUACUACGAUCUAAAGUUCUAUCACAAUAAGUUGUGGUGAAG- ((((((((.((((((............)))))))))((((....)))))))))..- ( -13.60, z-score = -2.01, R) >droSim1.chr3L 8796818 55 - 22553184 UCACCGAUCAGGGCUACUACGAUCUAAAGUUCUAUCACAAUAAGUUGUGGUGAAG- ((((((((.((((((............)))))))))((((....)))))))))..- ( -13.60, z-score = -2.01, R) >triCas2.ChLG3 320128 53 - 32080666 UAACGGAGCAGGGAUUUUUCGAUUUGAAAUUUUAUCAUAAUAAAUUGUGGUAA--- ...(((((........)))))..........(((((((((....)))))))))--- ( -6.90, z-score = -0.78, R) >consensus UCACCGAUCAGGGCUACUACGAUCUAAAGUUCUAUCACAAUAAAUUGUGGUAAAG_ ...((.....))....................((((((((....)))))))).... ( -7.36 = -7.16 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:13:44 2011